You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000959_00444
You are here: Home > Sequence: MGYG000000959_00444
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900752785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900752785 | |||||||||||
| CAZyme ID | MGYG000000959_00444 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14914; End: 17547 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 44 | 444 | 1.2e-96 | 0.9976076555023924 |
| CBM79 | 663 | 770 | 6.1e-42 | 0.9636363636363636 |
| CBM79 | 506 | 613 | 1.8e-38 | 0.9727272727272728 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 3.65e-81 | 47 | 436 | 1 | 367 | Glycosyl hydrolase family 9. |
| pfam18522 | DUF5620 | 2.40e-34 | 664 | 773 | 1 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| PLN02420 | PLN02420 | 6.26e-33 | 27 | 436 | 24 | 495 | endoglucanase |
| PLN02909 | PLN02909 | 1.78e-32 | 11 | 430 | 1 | 467 | Endoglucanase |
| PLN02613 | PLN02613 | 1.26e-31 | 38 | 426 | 20 | 458 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EWM53237.1 | 3.55e-211 | 5 | 875 | 3 | 880 |
| CBL17554.1 | 2.88e-187 | 7 | 462 | 9 | 464 |
| CDE33541.1 | 2.03e-167 | 7 | 563 | 8 | 591 |
| CDF00679.1 | 2.33e-103 | 513 | 778 | 1 | 271 |
| ALX07412.1 | 1.25e-96 | 1 | 455 | 1 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YIK_A | 8.66e-98 | 34 | 455 | 29 | 521 | ChainA, Endoglucanase [Acetivibrio thermocellus] |
| 1IA6_A | 4.79e-78 | 40 | 445 | 1 | 425 | CrystalStructure Of The Cellulase Cel9m Of C. Cellulolyticum [Ruminiclostridium cellulolyticum],1IA7_A Crystal Structure Of The Cellulase Cel9m Of C. Cellulolyticium In Complex With Cellobiose [Ruminiclostridium cellulolyticum] |
| 2XFG_A | 3.43e-59 | 44 | 450 | 25 | 463 | ChainA, ENDOGLUCANASE 1 [Acetivibrio thermocellus] |
| 5GXX_A | 8.43e-57 | 40 | 450 | 2 | 429 | ChainA, Glucanase [Acetivibrio thermocellus],5GXX_B Chain B, Glucanase [Acetivibrio thermocellus],5GXY_A Chain A, Glucanase [Acetivibrio thermocellus],5GXY_B Chain B, Glucanase [Acetivibrio thermocellus],5GXZ_A Chain A, Glucanase [Acetivibrio thermocellus],5GXZ_B Chain B, Glucanase [Acetivibrio thermocellus] |
| 5GY0_A | 5.44e-56 | 40 | 450 | 2 | 429 | ChainA, Glucanase [Acetivibrio thermocellus],5GY0_B Chain B, Glucanase [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02934 | 4.25e-56 | 44 | 465 | 77 | 529 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| Q5YLG1 | 1.98e-53 | 41 | 453 | 45 | 489 | Endoglucanase A OS=Bacillus pumilus OX=1408 GN=eglA PE=1 SV=1 |
| P28622 | 1.62e-52 | 41 | 448 | 26 | 465 | Endoglucanase 4 OS=Bacillus sp. (strain KSM-522) OX=120046 PE=3 SV=2 |
| P26221 | 1.70e-52 | 23 | 450 | 35 | 490 | Endoglucanase E-4 OS=Thermobifida fusca OX=2021 GN=celD PE=1 SV=2 |
| P26224 | 1.97e-52 | 40 | 450 | 27 | 467 | Endoglucanase F OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
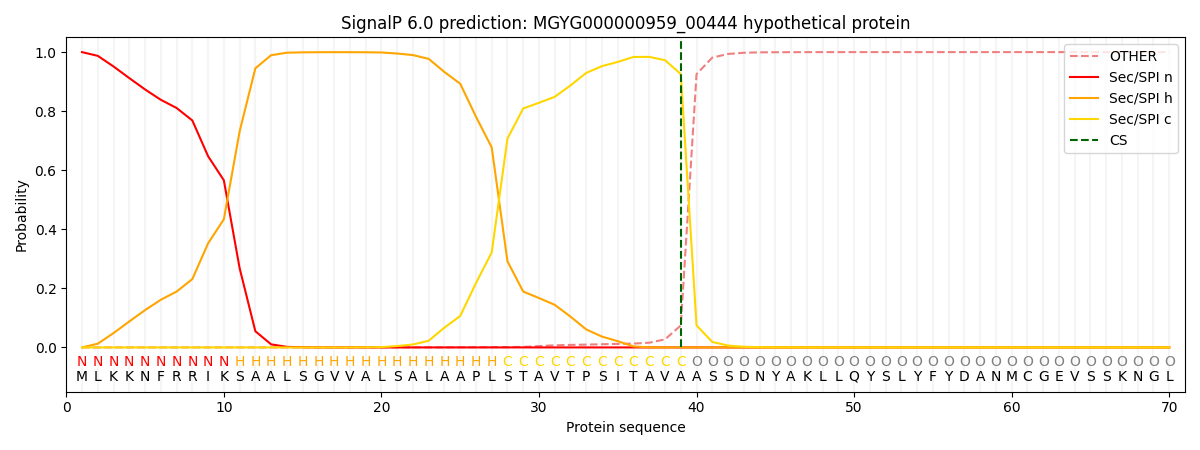
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000760 | 0.998078 | 0.000223 | 0.000430 | 0.000257 | 0.000210 |
