You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000959_01163
You are here: Home > Sequence: MGYG000000959_01163
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
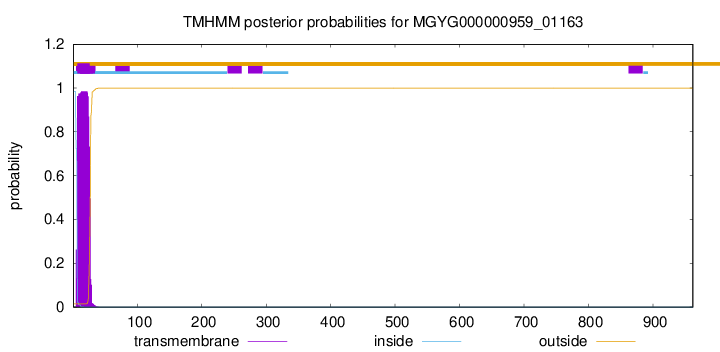
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900752785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900752785 | |||||||||||
| CAZyme ID | MGYG000000959_01163 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24842; End: 27730 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 264 | 448 | 1.1e-54 | 0.9076923076923077 |
| CBM13 | 563 | 704 | 1.4e-22 | 0.6808510638297872 |
| CBM13 | 717 | 874 | 6.8e-16 | 0.776595744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 9.76e-41 | 174 | 519 | 17 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 8.97e-21 | 262 | 445 | 30 | 208 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| smart00656 | Amb_all | 5.40e-20 | 260 | 448 | 10 | 186 | Amb_all domain. |
| pfam14200 | RicinB_lectin_2 | 7.54e-19 | 647 | 745 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 9.91e-19 | 601 | 690 | 5 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16867.1 | 0.0 | 37 | 938 | 35 | 899 |
| CDM68184.1 | 4.97e-119 | 38 | 620 | 29 | 584 |
| CDM70399.1 | 2.90e-96 | 38 | 703 | 31 | 694 |
| AKQ74328.1 | 1.96e-92 | 36 | 521 | 29 | 479 |
| QII50179.1 | 5.31e-92 | 36 | 521 | 29 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M2S_B | 1.41e-16 | 889 | 957 | 25 | 101 | R.flavefaciens' third ScaB cohesin in complex with a group 1 dockerin [Ruminococcus flavefaciens FD-1] |
| 5M2O_B | 2.16e-16 | 893 | 954 | 29 | 90 | R.flavefaciens' third ScaB cohesin in complex with a group 1 dockerin [Ruminococcus flavefaciens FD-1] |
| 3ZSC_A | 8.90e-13 | 200 | 425 | 14 | 212 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 5N5P_B | 9.63e-07 | 891 | 930 | 7 | 46 | Crystalstructure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A [Ruminococcus flavefaciens FD-1],5N5P_D Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A [Ruminococcus flavefaciens FD-1] |
| 5B2H_A | 6.01e-06 | 631 | 755 | 163 | 278 | Crystalstructure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum],5B2H_B Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94449 | 7.15e-58 | 184 | 522 | 32 | 338 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| O34819 | 1.34e-57 | 184 | 522 | 32 | 338 | Pectin lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pelB PE=3 SV=1 |
| P27027 | 4.46e-48 | 187 | 521 | 10 | 304 | Pectin lyase OS=Pseudomonas marginalis OX=298 GN=pnl PE=1 SV=2 |
| P24112 | 2.71e-42 | 184 | 523 | 7 | 308 | Pectin lyase OS=Pectobacterium carotovorum OX=554 GN=pnl PE=1 SV=1 |
| Q9WYR4 | 6.08e-13 | 200 | 425 | 41 | 239 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
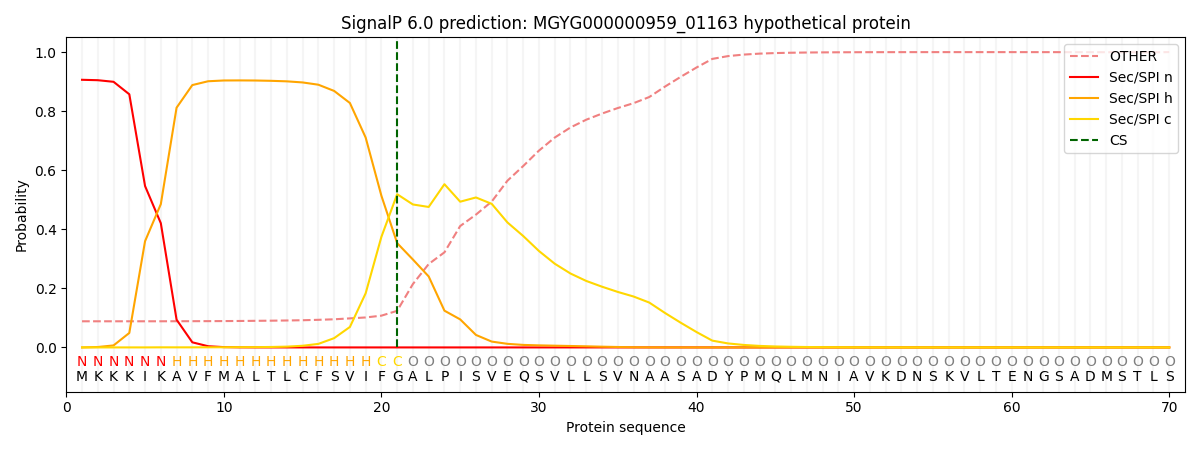
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.095818 | 0.897103 | 0.006160 | 0.000269 | 0.000280 | 0.000342 |