You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000964_01380
You are here: Home > Sequence: MGYG000000964_01380
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola salanitronis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola salanitronis | |||||||||||
| CAZyme ID | MGYG000000964_01380 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16255; End: 19122 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 17 | 662 | 1.4e-83 | 0.6356382978723404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.96e-30 | 23 | 478 | 13 | 433 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.97e-25 | 17 | 473 | 7 | 443 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 3.53e-16 | 25 | 218 | 4 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK09525 | lacZ | 1.16e-14 | 97 | 401 | 103 | 410 | beta-galactosidase. |
| PRK10340 | ebgA | 1.07e-13 | 100 | 384 | 95 | 377 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37834.1 | 0.0 | 1 | 955 | 1 | 955 |
| QUT56820.1 | 0.0 | 1 | 954 | 1 | 954 |
| QEW35558.1 | 0.0 | 1 | 954 | 1 | 954 |
| QQY39766.1 | 0.0 | 1 | 954 | 1 | 954 |
| QQY43067.1 | 0.0 | 1 | 954 | 1 | 954 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BGA_A | 7.08e-20 | 14 | 450 | 40 | 464 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6U7I_A | 1.51e-19 | 105 | 466 | 57 | 426 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 3DEC_A | 6.27e-19 | 14 | 450 | 36 | 460 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 7VQM_A | 1.90e-18 | 109 | 450 | 91 | 409 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 6ZJV_A | 9.70e-18 | 143 | 473 | 157 | 462 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P24131 | 1.83e-18 | 24 | 474 | 48 | 481 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
| Q9K9C6 | 4.05e-17 | 116 | 474 | 124 | 482 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q04F24 | 2.77e-16 | 20 | 450 | 44 | 457 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| Q59140 | 2.44e-15 | 97 | 450 | 97 | 434 | Beta-galactosidase OS=Arthrobacter sp. (strain B7) OX=86041 GN=lacZ PE=1 SV=1 |
| P23989 | 7.30e-15 | 25 | 473 | 48 | 479 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
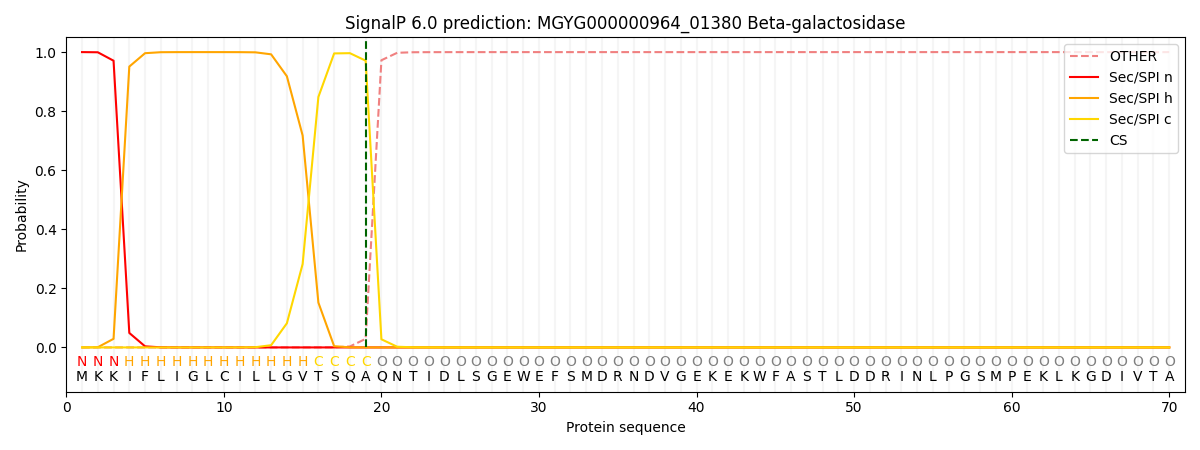
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000588 | 0.998212 | 0.000415 | 0.000261 | 0.000261 | 0.000238 |
