You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000964_01846
You are here: Home > Sequence: MGYG000000964_01846
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola salanitronis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola salanitronis | |||||||||||
| CAZyme ID | MGYG000000964_01846 | |||||||||||
| CAZy Family | PL26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11874; End: 15110 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL26 | 241 | 1076 | 0 | 0.9918793503480279 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37515.1 | 0.0 | 1 | 1078 | 1 | 1078 |
| VTZ55478.1 | 0.0 | 22 | 1078 | 11 | 1075 |
| QQY43085.1 | 0.0 | 1 | 1077 | 1 | 1094 |
| QUT56846.1 | 0.0 | 1 | 1077 | 8 | 1101 |
| ABR37887.1 | 0.0 | 1 | 1077 | 8 | 1101 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XQG_A | 6.41e-164 | 238 | 1070 | 7 | 891 | Crystalstructure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_B Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_C Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_D Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_E Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_F Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_G Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQG_H Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose [Penicillium chrysogenum],5XQJ_A Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_B Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_C Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_D Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_E Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_F Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_G Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum],5XQJ_H Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose [Penicillium chrysogenum] |
| 5XQ3_B | 2.67e-162 | 238 | 1070 | 7 | 891 | Crystalstructure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum [Penicillium chrysogenum] |
| 5XQ3_A | 2.82e-162 | 238 | 1070 | 7 | 891 | Crystalstructure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum [Penicillium chrysogenum] |
| 5XQO_A | 1.04e-161 | 238 | 1070 | 7 | 891 | Crystalstructure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate [Penicillium chrysogenum],5XQO_B Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate [Penicillium chrysogenum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31530 | 5.85e-189 | 241 | 1076 | 4 | 842 | Uncharacterized protein YetA OS=Bacillus subtilis (strain 168) OX=224308 GN=yetA PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
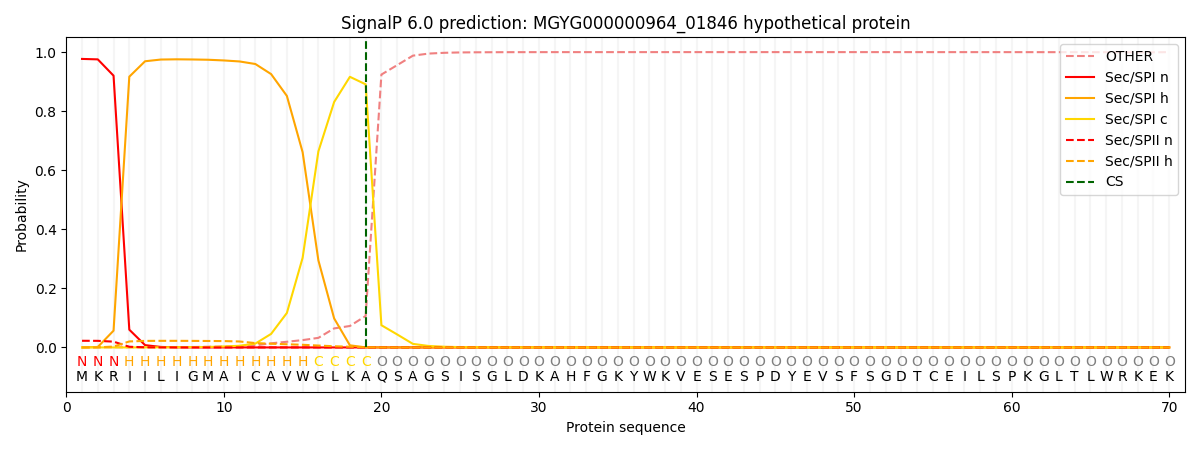
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002056 | 0.972620 | 0.024491 | 0.000307 | 0.000259 | 0.000241 |
