You are browsing environment: HUMAN GUT
MGYG000000964_02362
Basic Information
help
Species
Phocaeicola salanitronis
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola salanitronis
CAZyme ID
MGYG000000964_02362
CAZy Family
GH28
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000964
3100012
MAG
Denmark
Europe
Gene Location
Start: 6956;
End: 8389
Strand: -
No EC number prediction in MGYG000000964_02362.
Family
Start
End
Evalue
family coverage
GH28
174
443
8.5e-32
0.7876923076923077
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00295
Glyco_hydro_28
0.009
261
410
139
274
Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
B0Y9F8
6.57e-06
179
401
99
337
Probable endo-xylogalacturonan hydrolase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xghA PE=3 SV=2
more
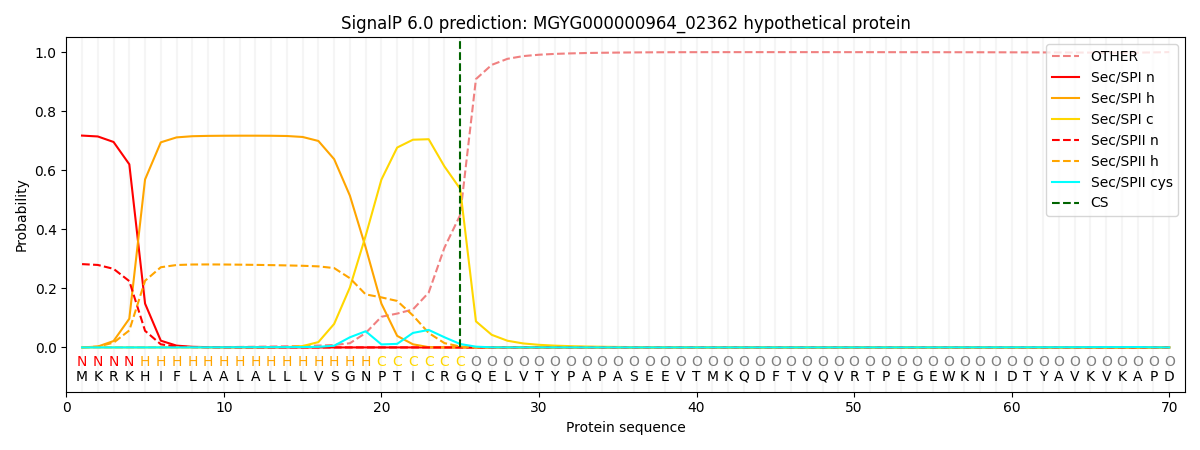
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.001456
0.708292
0.289322
0.000450
0.000237
0.000208
There is no transmembrane helices in MGYG000000964_02362.