You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000965_01244
You are here: Home > Sequence: MGYG000000965_01244
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | PeH17 sp900542285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-138; PeH17; PeH17 sp900542285 | |||||||||||
| CAZyme ID | MGYG000000965_01244 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10523; End: 13300 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 445 | 675 | 4.2e-52 | 0.905829596412556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 8.71e-27 | 727 | 912 | 7 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 6.90e-19 | 724 | 916 | 502 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam02018 | CBM_4_9 | 0.001 | 32 | 150 | 2 | 123 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam13231 | PMT_2 | 0.001 | 491 | 610 | 22 | 133 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| pfam15425 | DUF4627 | 0.001 | 35 | 110 | 2 | 90 | Domain of unknown function (DUF4627). This family of proteins is found in bacteria. Proteins in this family are approximately 230 amino acids in length. There is a conserved WYK sequence motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUG58294.1 | 3.32e-123 | 4 | 916 | 2 | 960 |
| AEV69574.1 | 3.93e-121 | 10 | 909 | 5 | 859 |
| ADU73726.1 | 3.11e-120 | 6 | 916 | 3 | 968 |
| ANV75398.1 | 3.11e-120 | 6 | 916 | 3 | 968 |
| ALX07656.1 | 3.11e-120 | 6 | 916 | 3 | 968 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L8F4Z2 | 1.41e-21 | 448 | 914 | 67 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| P9WN04 | 4.58e-21 | 448 | 914 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 4.58e-21 | 448 | 914 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q8NRZ6 | 4.64e-15 | 471 | 914 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
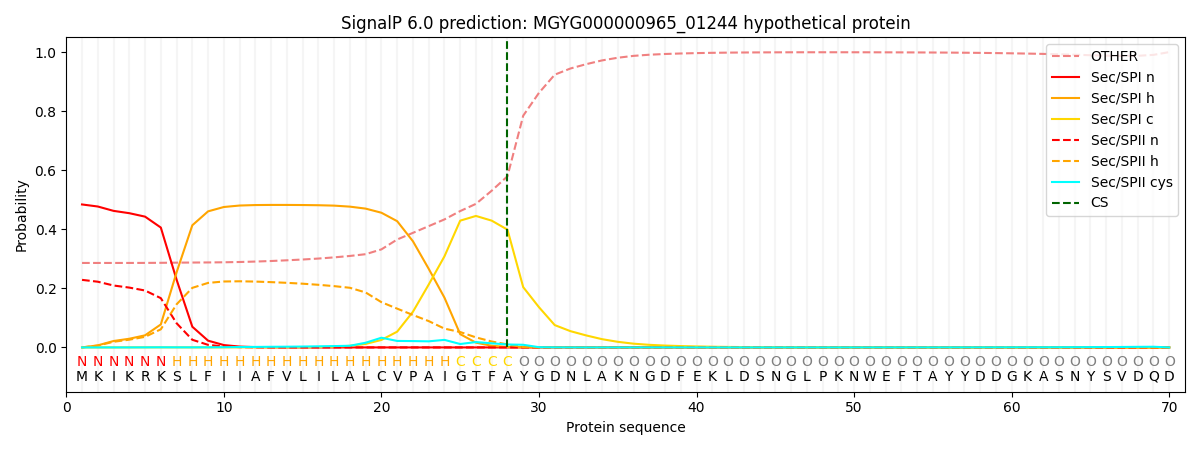
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.300372 | 0.457081 | 0.235237 | 0.003479 | 0.001522 | 0.002293 |
TMHMM Annotations download full data without filtering help
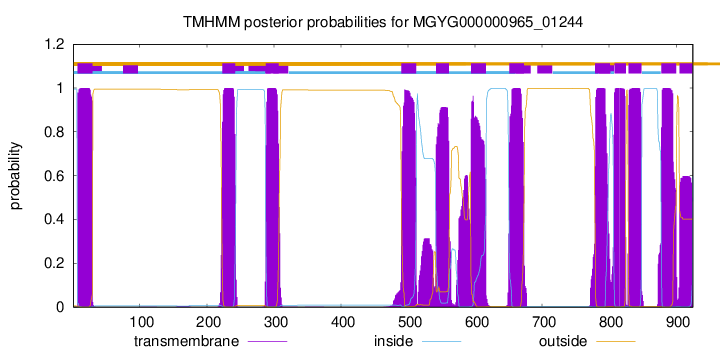
| start | end |
|---|---|
| 7 | 29 |
| 223 | 242 |
| 288 | 307 |
| 490 | 512 |
| 542 | 561 |
| 594 | 616 |
| 651 | 673 |
| 779 | 801 |
| 808 | 825 |
| 829 | 848 |
| 878 | 900 |
| 905 | 924 |
