You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000967_01219
You are here: Home > Sequence: MGYG000000967_01219
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
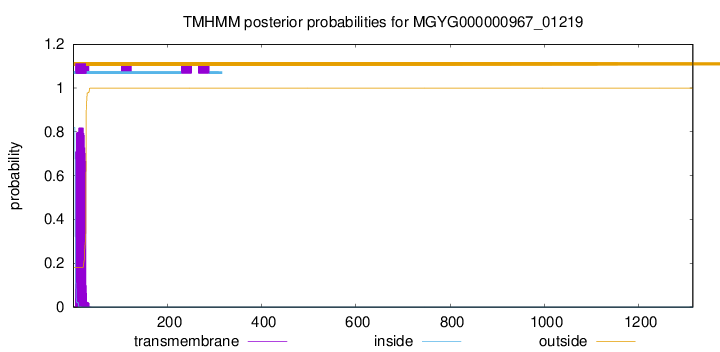
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900543365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900543365 | |||||||||||
| CAZyme ID | MGYG000000967_01219 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 553; End: 4500 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 949 | 1289 | 6.8e-105 | 0.8986666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07523 | Big_3 | 2.25e-04 | 499 | 565 | 2 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| cd14251 | PL-6 | 8.16e-04 | 973 | 1103 | 9 | 133 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam07523 | Big_3 | 0.002 | 246 | 307 | 4 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 0.002 | 413 | 486 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam14592 | Chondroitinas_B | 0.005 | 966 | 1070 | 2 | 107 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAC87940.1 | 1.09e-214 | 63 | 1264 | 70 | 1193 |
| AGI38277.1 | 1.53e-214 | 63 | 1264 | 70 | 1193 |
| AGC67199.1 | 1.53e-214 | 63 | 1264 | 70 | 1193 |
| ANW97669.1 | 1.53e-214 | 63 | 1264 | 70 | 1193 |
| ANX00231.1 | 2.15e-214 | 63 | 1264 | 70 | 1193 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 5.27e-32 | 952 | 1233 | 18 | 292 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 4.45e-09 | 1096 | 1201 | 168 | 275 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 4.78e-66 | 573 | 1259 | 34 | 663 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 5.55e-32 | 952 | 1233 | 43 | 317 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 4.47e-31 | 952 | 1233 | 43 | 317 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| A0A401ETL2 | 1.00e-06 | 319 | 486 | 1301 | 1451 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
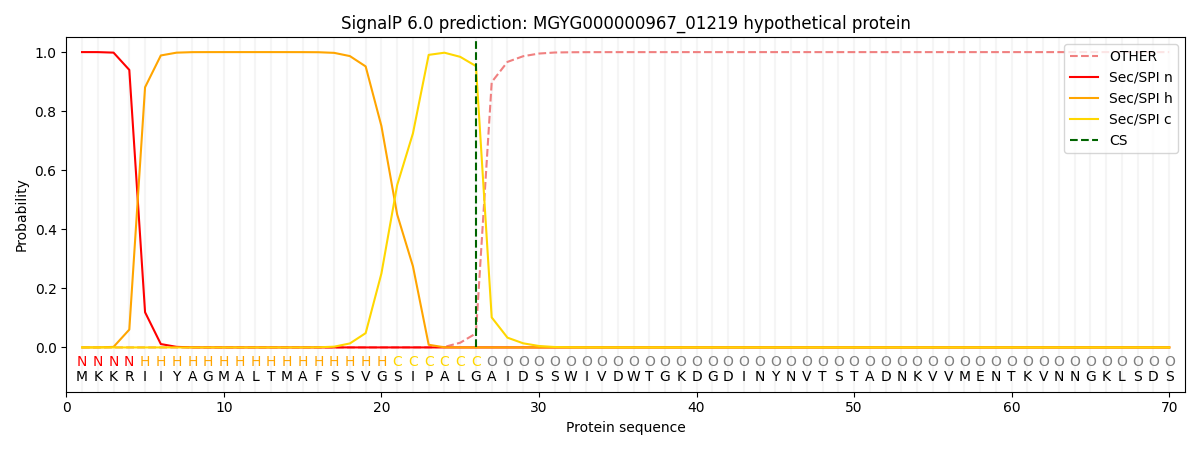
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000277 | 0.999101 | 0.000155 | 0.000170 | 0.000147 | 0.000136 |