You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000971_01057
You are here: Home > Sequence: MGYG000000971_01057
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
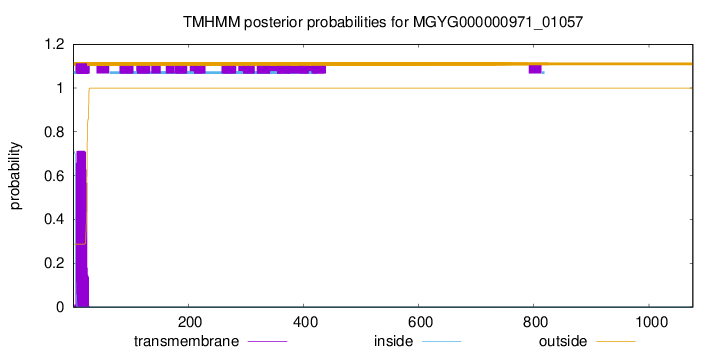
TMHMM annotations
Basic Information help
| Species | UMGS1518 sp900552575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; UMGS1518; UMGS1518 sp900552575 | |||||||||||
| CAZyme ID | MGYG000000971_01057 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8725; End: 11955 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 37 | 678 | 2.4e-45 | 0.6781914893617021 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 5.75e-12 | 237 | 436 | 222 | 408 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 8.13e-10 | 259 | 436 | 278 | 451 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAX78846.1 | 1.36e-255 | 40 | 1073 | 29 | 1080 |
| QTE22603.1 | 2.66e-241 | 44 | 1071 | 24 | 1062 |
| QOD61956.1 | 5.90e-240 | 44 | 1071 | 24 | 1062 |
| QQT78362.1 | 2.23e-235 | 38 | 1073 | 46 | 1094 |
| QRP56393.1 | 2.23e-235 | 38 | 1073 | 46 | 1094 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NCW_A | 1.38e-10 | 78 | 437 | 35 | 381 | Crystalstructure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_B Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_C Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_D Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCX_A Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_B Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_C Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_D Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi] |
| 6D89_A | 1.36e-07 | 262 | 462 | 259 | 443 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 6D1N_A | 1.38e-07 | 262 | 462 | 267 | 451 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
| 6XXW_A | 1.61e-06 | 264 | 456 | 250 | 437 | Structureof beta-D-Glucuronidase for Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
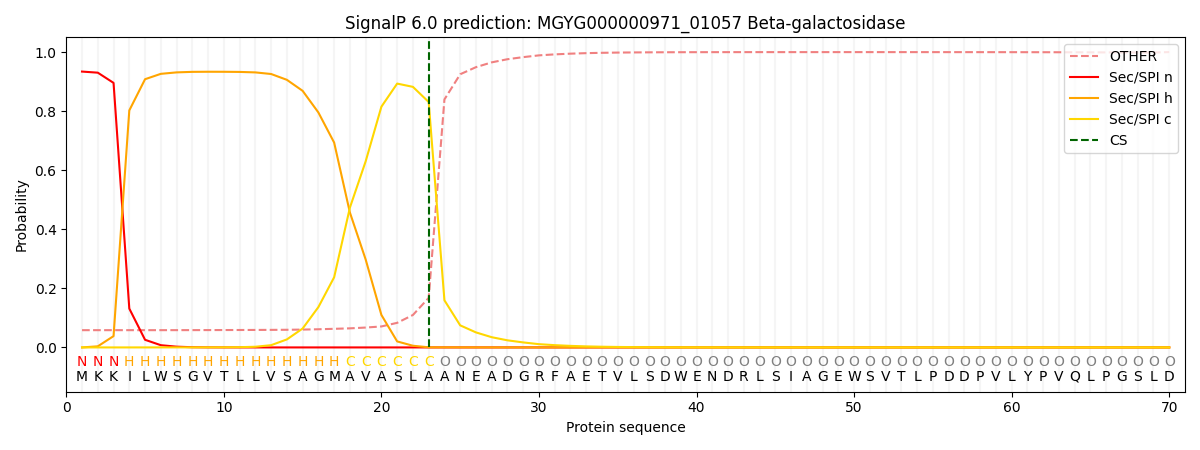
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.068622 | 0.921666 | 0.008365 | 0.000591 | 0.000352 | 0.000368 |