You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000977_00789
You are here: Home > Sequence: MGYG000000977_00789
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium sp900539375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900539375 | |||||||||||
| CAZyme ID | MGYG000000977_00789 | |||||||||||
| CAZy Family | CBM74 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55976; End: 58894 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM74 | 51 | 342 | 1.6e-76 | 0.9935897435897436 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033187 | internalin_J | 2.74e-48 | 308 | 694 | 457 | 815 | class 1 internalin InlJ. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin J (InlJ). |
| PRK14081 | PRK14081 | 3.60e-24 | 580 | 849 | 105 | 378 | triple tyrosine motif-containing protein; Provisional |
| PRK14081 | PRK14081 | 2.12e-20 | 602 | 834 | 222 | 458 | triple tyrosine motif-containing protein; Provisional |
| pfam06458 | MucBP | 4.50e-19 | 433 | 491 | 2 | 60 | MucBP domain. The MucBP (MUCin-Binding Protein) domain is found in a wide variety of bacterial proteins, in several repeats. The domain is found in bacterial peptidoglycan bound proteins and is often found in conjunction with pfam00746 and pfam00560. |
| pfam06458 | MucBP | 1.46e-18 | 504 | 564 | 1 | 61 | MucBP domain. The MucBP (MUCin-Binding Protein) domain is found in a wide variety of bacterial proteins, in several repeats. The domain is found in bacterial peptidoglycan bound proteins and is often found in conjunction with pfam00746 and pfam00560. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDM67953.1 | 7.57e-138 | 49 | 971 | 776 | 1583 |
| QCT07491.1 | 1.02e-128 | 43 | 352 | 218 | 533 |
| AWX97480.1 | 4.42e-114 | 35 | 447 | 739 | 1154 |
| AWX95531.1 | 4.42e-114 | 35 | 447 | 739 | 1154 |
| QZS61962.1 | 1.18e-104 | 77 | 447 | 1 | 368 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KT7_A | 1.10e-09 | 425 | 515 | 2 | 81 | ChainA, Putative peptidoglycan bound protein (LPXTG motif) [Listeria monocytogenes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q71VU0 | 1.37e-46 | 355 | 577 | 577 | 797 | Internalin J OS=Listeria monocytogenes serotype 4b (strain F2365) OX=265669 GN=inlJ PE=3 SV=1 |
| Q8Y3L4 | 1.64e-43 | 308 | 577 | 457 | 727 | Internalin J OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlJ PE=1 SV=1 |
| Q723X5 | 4.40e-22 | 360 | 569 | 1513 | 1712 | Internalin I OS=Listeria monocytogenes serotype 4b (strain F2365) OX=265669 GN=inlI PE=3 SV=1 |
| Q8YA32 | 2.25e-21 | 360 | 569 | 1516 | 1715 | Internalin I OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlI PE=2 SV=1 |
| P32653 | 7.59e-06 | 360 | 536 | 958 | 1129 | Muramidase-released protein OS=Streptococcus suis OX=1307 GN=mrp PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
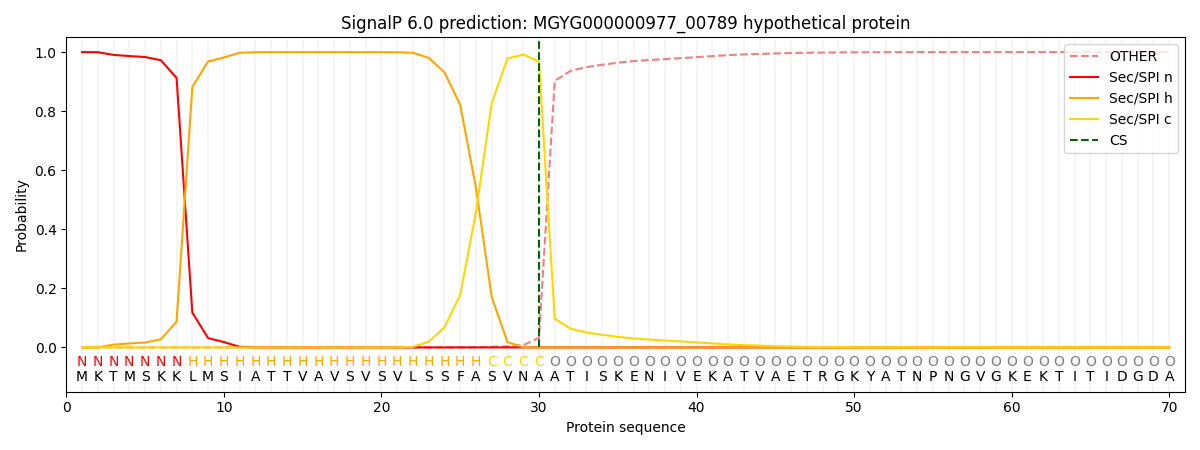
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000343 | 0.998857 | 0.000248 | 0.000195 | 0.000175 | 0.000152 |
