You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000977_02116
You are here: Home > Sequence: MGYG000000977_02116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium sp900539375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium sp900539375 | |||||||||||
| CAZyme ID | MGYG000000977_02116 | |||||||||||
| CAZy Family | CBM9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14307; End: 18995 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM9 | 55 | 240 | 5.8e-28 | 0.9945054945054945 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08563 | GDPD_TtGDE_like | 3.12e-58 | 1047 | 1278 | 3 | 229 | Glycerophosphodiester phosphodiesterase domain of Thermoanaerobacter tengcongensis and similar proteins. This subfamily corresponds to the glycerophosphodiester phosphodiesterase domain (GDPD) present in Thermoanaerobacter tengcongensis glycerophosphodiester phosphodiesterase (TtGDE, EC 3.1.4.46) and its uncharacterized homologs. Members in this family show high sequence similarity to Escherichia coli GP-GDE, which catalyzes the degradation of glycerophosphodiesters to produce sn-glycerol-3-phosphate (G3P) and the corresponding alcohols. Despite the fact that most of GDPD family members exist as the monomer, TtGDE can function as a dimeric unit. Its catalytic mechanism is based on the general base-acid catalysis, which is similar to that of phosphoinositide-specific phospholipases C (PI-PLCs, EC 3.1.4.11). A divalent metal cation is required for the enzyme activity of TtGDE. |
| cd08582 | GDPD_like_2 | 3.85e-53 | 1047 | 1281 | 1 | 233 | Glycerophosphodiester phosphodiesterase domain of uncharacterized bacterial glycerophosphodiester phosphodiesterases. This subfamily corresponds to the glycerophosphodiester phosphodiesterase domain (GDPD) present in a group of uncharacterized bacterial glycerophosphodiester phosphodiesterase and similar proteins. They show high sequence similarity to Escherichia coli glycerophosphodiester phosphodiesterase, which catalyzes the degradation of glycerophosphodiesters to produce sn-glycerol-3-phosphate (G3P) and the corresponding alcohols. |
| cd08556 | GDPD | 2.88e-46 | 1047 | 1278 | 1 | 189 | Glycerophosphodiester phosphodiesterase domain as found in prokaryota and eukaryota, and similar proteins. The typical glycerophosphodiester phosphodiesterase domain (GDPD) consists of a TIM barrel and a small insertion domain named the GDPD-insertion (GDPD-I) domain, which is specific for GDPD proteins. This family corresponds to both typical GDPD domain and GDPD-like domain which lacks the GDPD-I region. Members in this family mainly consist of a large family of prokaryotic and eukaryotic glycerophosphodiester phosphodiesterases (GP-GDEs, EC 3.1.4.46), and a number of uncharacterized homologs. Sphingomyelinases D (SMases D) (sphingomyelin phosphodiesterase D, EC 3.1.4.41) from spider venom, SMases D-like proteins, and phospholipase D (PLD) from several pathogenic bacteria are also included in this family. GDPD plays an essential role in glycerol metabolism and catalyzes the hydrolysis of glycerophosphodiesters to sn-glycerol-3-phosphate (G3P) and the corresponding alcohols are major sources of carbon and phosphate. Its catalytic mechanism is based on the metal ion-dependent acid-base reaction, which is similar to that of phosphoinositide-specific phospholipases C (PI-PLCs, EC 3.1.4.11). Both, GDPD related proteins and PI-PLCs, belong to the superfamily of PI-PLC-like phosphodiesterases. |
| COG0584 | UgpQ | 8.24e-45 | 1043 | 1278 | 4 | 246 | Glycerophosphoryl diester phosphodiesterase [Lipid transport and metabolism]. |
| cd08601 | GDPD_SaGlpQ_like | 3.66e-43 | 1045 | 1280 | 1 | 249 | Glycerophosphodiester phosphodiesterase domain of Staphylococcus aureus and similar proteins. This subfamily corresponds to the glycerophosphodiester phosphodiesterase domain (GDPD) present in uncharacterized glycerophosphodiester phosphodiesterase (GP-GDE, EC 3.1.4.46) from Staphylococcus aureus, Bacillus subtilis and similar proteins. Members in this family show very high sequence similarity to Escherichia coli periplasmic phosphodiesterase GlpQ, which catalyzes the Ca2+-dependent degradation of periplasmic glycerophosphodiesters to produce sn-glycerol-3-phosphate (G3P) and the corresponding alcohols. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV16877.1 | 0.0 | 41 | 1375 | 37 | 1385 |
| ALS30162.1 | 0.0 | 40 | 1375 | 41 | 1396 |
| QJD84974.1 | 0.0 | 41 | 1381 | 37 | 1396 |
| QKS47384.1 | 0.0 | 38 | 1383 | 118 | 1479 |
| QCT04000.1 | 0.0 | 14 | 1375 | 12 | 1393 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2PZ0_A | 4.39e-35 | 1047 | 1276 | 14 | 238 | ChainA, Glycerophosphoryl diester phosphodiesterase [Caldanaerobacter subterraneus subsp. tengcongensis MB4],2PZ0_B Chain B, Glycerophosphoryl diester phosphodiesterase [Caldanaerobacter subterraneus subsp. tengcongensis MB4] |
| 5T91_A | 6.42e-29 | 1048 | 1278 | 16 | 257 | Crystalstructure of B. subtilis 168 GlpQ in complex with bicine [Bacillus subtilis subsp. subtilis str. 168],5T9B_G Crystal structure of B. subtilis 168 GlpQ in complex with glycerol-3-phosphate (5 minute soak) [Bacillus subtilis subsp. subtilis str. 168],5T9C_E Crystal structure of B. subtilis 168 GlpQ in complex with glycerol-3-phosphate (1 hour soak) [Bacillus subtilis subsp. subtilis str. 168] |
| 4R7O_A | 1.01e-24 | 1048 | 1278 | 19 | 281 | CrystalStructure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_B Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_C Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_D Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_E Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_F Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames],4R7O_G Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci [Bacillus anthracis str. Ames] |
| 2OOG_A | 8.57e-23 | 1048 | 1288 | 27 | 280 | ChainA, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2OOG_B Chain B, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2OOG_C Chain C, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2OOG_D Chain D, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2OOG_E Chain E, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2OOG_F Chain F, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_A Chain A, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_B Chain B, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_C Chain C, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_D Chain D, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_E Chain E, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_F Chain F, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_G Chain G, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315],2P76_H Chain H, Glycerophosphoryl diester phosphodiesterase [Staphylococcus aureus subsp. aureus N315] |
| 4OEC_A | 2.52e-19 | 1047 | 1202 | 9 | 153 | Crystalstructure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 [Thermococcus kodakarensis KOD1],4OEC_B Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 [Thermococcus kodakarensis KOD1],4OEC_C Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 [Thermococcus kodakarensis KOD1],4OEC_D Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 [Thermococcus kodakarensis KOD1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O07592 | 4.74e-28 | 1046 | 1278 | 2 | 230 | Putative glycerophosphodiester phosphodiesterase YhdW OS=Bacillus subtilis (strain 168) OX=224308 GN=yhdW PE=3 SV=1 |
| P37965 | 6.31e-28 | 1048 | 1278 | 41 | 282 | Glycerophosphodiester phosphodiesterase OS=Bacillus subtilis (strain 168) OX=224308 GN=glpQ PE=1 SV=1 |
| Q9NZC3 | 4.36e-22 | 1048 | 1192 | 68 | 215 | Glycerophosphodiester phosphodiesterase 1 OS=Homo sapiens OX=9606 GN=GDE1 PE=1 SV=1 |
| Q3T0T0 | 2.12e-20 | 1048 | 1192 | 68 | 215 | Glycerophosphodiester phosphodiesterase 1 OS=Bos taurus OX=9913 GN=GDE1 PE=2 SV=1 |
| Q9JL55 | 9.42e-20 | 1048 | 1192 | 68 | 215 | Glycerophosphodiester phosphodiesterase 1 OS=Rattus norvegicus OX=10116 GN=Gde1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
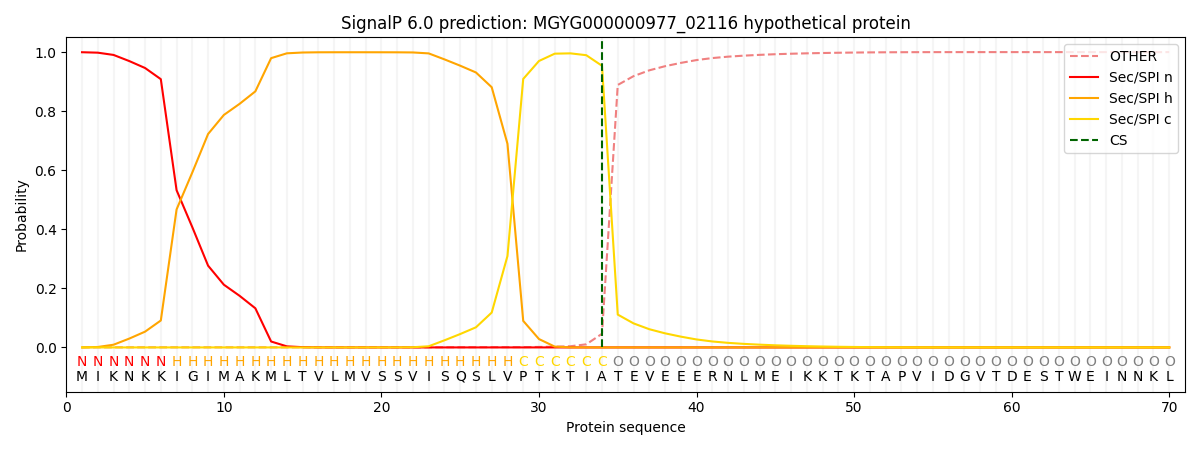
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001002 | 0.998065 | 0.000346 | 0.000215 | 0.000172 | 0.000159 |
