You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000978_00513
You are here: Home > Sequence: MGYG000000978_00513
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
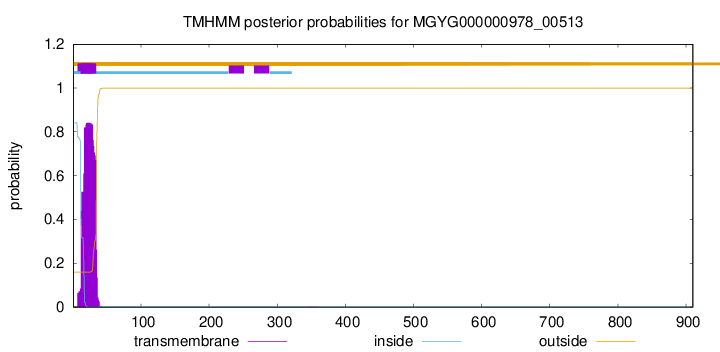
TMHMM annotations
Basic Information help
| Species | Streptococcus mitis_D | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus mitis_D | |||||||||||
| CAZyme ID | MGYG000000978_00513 | |||||||||||
| CAZy Family | CBM47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19322; End: 22054 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM47 | 515 | 634 | 6.7e-35 | 0.984375 |
| CBM47 | 649 | 770 | 6.5e-32 | 0.984375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17440 | Thiol_cytolys_C | 4.07e-31 | 802 | 903 | 1 | 102 | Thiol-activated cytolysin beta sandwich domain. This domain has an immunoglobulin like fold. It is found at the C-terminus of the thiol-activated cytolsin protein. |
| smart00607 | FTP | 2.34e-13 | 505 | 579 | 1 | 80 | eel-Fucolectin Tachylectin-4 Pentaxrin-1 Domain. |
| smart00607 | FTP | 9.05e-12 | 639 | 773 | 1 | 145 | eel-Fucolectin Tachylectin-4 Pentaxrin-1 Domain. |
| cd00741 | Lipase | 3.06e-08 | 325 | 448 | 10 | 108 | Lipase. Lipases are esterases that can hydrolyze long-chain acyl-triglycerides into di- and monoglycerides, glycerol, and free fatty acids at a water/lipid interface. A typical feature of lipases is "interfacial activation", the process of becoming active at the lipid/water interface, although several examples of lipases have been identified that do not undergo interfacial activation . The active site of a lipase contains a catalytic triad consisting of Ser - His - Asp/Glu, but unlike most serine proteases, the active site is buried inside the structure. A "lid" or "flap" covers the active site, making it inaccessible to solvent and substrates. The lid opens during the process of interfacial activation, allowing the lipid substrate access to the active site. |
| pfam00754 | F5_F8_type_C | 2.58e-07 | 514 | 627 | 2 | 116 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEL10282.1 | 0.0 | 1 | 910 | 1 | 915 |
| BCJ10480.1 | 0.0 | 60 | 729 | 5 | 677 |
| VFI04050.1 | 5.68e-290 | 49 | 595 | 1 | 546 |
| VFI27642.1 | 5.68e-290 | 49 | 595 | 1 | 546 |
| AJD72412.1 | 6.49e-199 | 266 | 595 | 1 | 330 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3LE0_A | 1.58e-42 | 640 | 778 | 10 | 149 | ChainA, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEG_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEI_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis],3LEK_A Chain A, Platelet aggregation factor Sm-hPAF [Streptococcus mitis] |
| 4GWI_A | 5.50e-42 | 640 | 778 | 10 | 149 | His62 mutant of the lectin binding domain of lectinolysin complexed with Lewis y [Streptococcus mitis],4GWJ_A His 62 mutant of the lectin binding domain of Lectinolysin complexed with Lewis b [Streptococcus mitis] |
| 2J1R_A | 4.44e-36 | 641 | 774 | 17 | 149 | Structureof a Streptococcus pneumoniae fucose binding module [Streptococcus pneumoniae TIGR4],2J1R_B Structure of a Streptococcus pneumoniae fucose binding module [Streptococcus pneumoniae TIGR4],2J1S_A Structure of a Streptococcus pneumoniae fucose binding module in complex with fucose [Streptococcus pneumoniae TIGR4],2J1S_B Structure of a Streptococcus pneumoniae fucose binding module in complex with fucose [Streptococcus pneumoniae TIGR4],2J1T_A Structure of a Streptococcus pneumoniae fucose binding module in complex with the Lewis Y antigen [Streptococcus pneumoniae TIGR4],2J1T_B Structure of a Streptococcus pneumoniae fucose binding module in complex with the Lewis Y antigen [Streptococcus pneumoniae TIGR4],2J1U_A Structure of a Streptococcus pneumoniae fucose binding module in complex with the blood group A-tetrasaccharide [Streptococcus pneumoniae TIGR4],2J1U_B Structure of a Streptococcus pneumoniae fucose binding module in complex with the blood group A-tetrasaccharide [Streptococcus pneumoniae TIGR4],2J1V_A Structure of a Streptococcus pneumoniae fucose binding module in complex with the blood group H-trisaccharide [Streptococcus pneumoniae TIGR4],2J1V_B Structure of a Streptococcus pneumoniae fucose binding module in complex with the blood group H-trisaccharide [Streptococcus pneumoniae TIGR4] |
| 2J22_A | 1.47e-33 | 638 | 775 | 11 | 147 | Structureof a Streptococcus pneumoniae fucose binding module, SpX-3 [Streptococcus pneumoniae TIGR4] |
| 3CQO_A | 4.69e-33 | 506 | 773 | 4 | 288 | Crystalstructure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis],3CQO_B Crystal structure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis],3CQO_C Crystal structure of a f-lectin (fucolectin) from morone saxatilis (striped bass) serum [Morone saxatilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q04IN8 | 2.27e-22 | 775 | 906 | 335 | 466 | Pneumolysin OS=Streptococcus pneumoniae serotype 2 (strain D39 / NCTC 7466) OX=373153 GN=ply PE=1 SV=1 |
| Q7ZAK5 | 2.27e-22 | 775 | 906 | 335 | 466 | Pneumolysin OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=ply PE=3 SV=1 |
| P0C2J9 | 4.05e-22 | 775 | 906 | 335 | 466 | Pneumolysin OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=ply PE=1 SV=1 |
| Q893D9 | 4.46e-20 | 803 | 905 | 420 | 522 | Tetanolysin OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=CTC_01888 PE=1 SV=1 |
| P31831 | 2.36e-18 | 800 | 906 | 415 | 521 | Ivanolysin OS=Listeria ivanovii OX=1638 GN=ilo PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
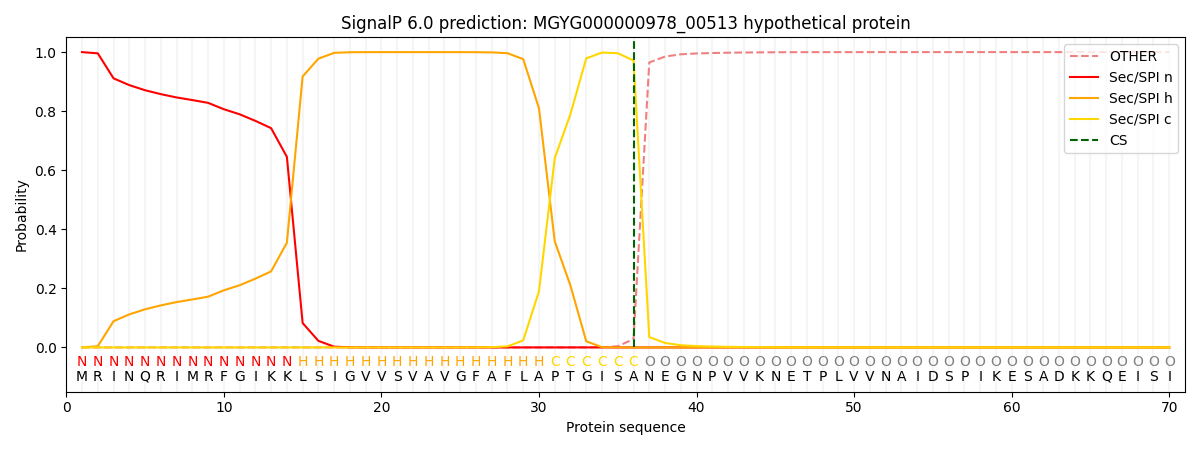
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000370 | 0.998901 | 0.000198 | 0.000182 | 0.000173 | 0.000149 |