You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000988_00784
You are here: Home > Sequence: MGYG000000988_00784
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
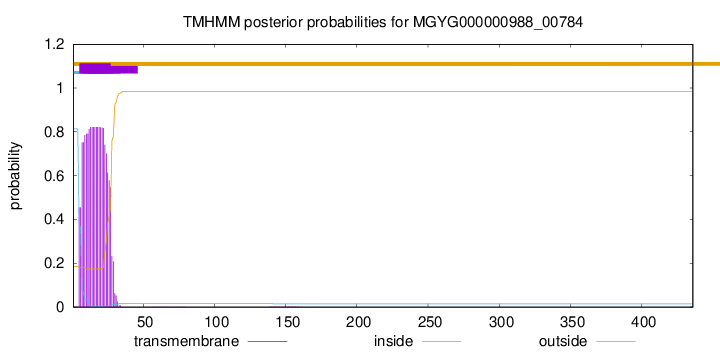
TMHMM annotations
Basic Information help
| Species | Anaerostipes rhamnosivorans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerostipes; Anaerostipes rhamnosivorans | |||||||||||
| CAZyme ID | MGYG000000988_00784 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33068; End: 34378 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 256 | 416 | 7.9e-26 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 4.90e-37 | 174 | 436 | 19 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 3.41e-13 | 245 | 417 | 4 | 142 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam07833 | Cu_amine_oxidN1 | 2.36e-07 | 82 | 166 | 17 | 93 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
| pfam01832 | Glucosaminidase | 2.05e-04 | 260 | 364 | 7 | 76 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP36276.1 | 3.55e-315 | 1 | 436 | 1 | 436 |
| BCD36607.1 | 2.87e-289 | 1 | 436 | 1 | 436 |
| QMW70532.1 | 2.87e-289 | 1 | 436 | 1 | 436 |
| AQP40018.1 | 4.75e-176 | 22 | 435 | 20 | 447 |
| CBL39631.1 | 9.55e-176 | 22 | 435 | 20 | 447 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WQW_A | 1.43e-45 | 181 | 435 | 21 | 269 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
| 6FXO_A | 4.48e-27 | 215 | 436 | 57 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 1.86e-20 | 238 | 418 | 68 | 215 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.22e-19 | 238 | 418 | 68 | 215 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXP_A | 1.48e-15 | 220 | 425 | 71 | 237 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 5.19e-25 | 213 | 436 | 697 | 880 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q6GI31 | 2.64e-24 | 215 | 436 | 1070 | 1257 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
| A7X0T9 | 2.64e-24 | 215 | 436 | 1068 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
| Q99V41 | 2.64e-24 | 215 | 436 | 1061 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 2.64e-24 | 215 | 436 | 1061 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
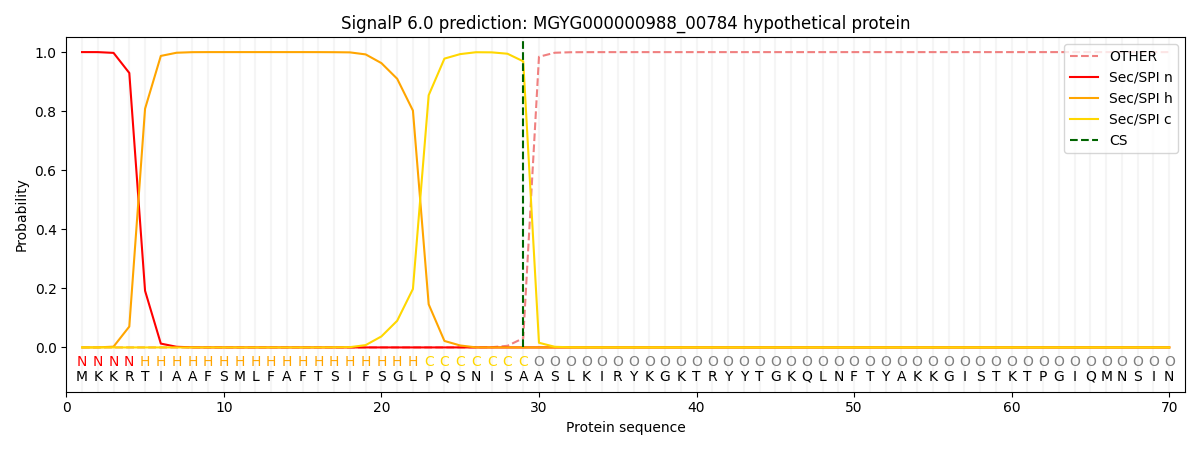
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000291 | 0.999016 | 0.000200 | 0.000179 | 0.000164 | 0.000146 |