You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000989_03048
You are here: Home > Sequence: MGYG000000989_03048
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
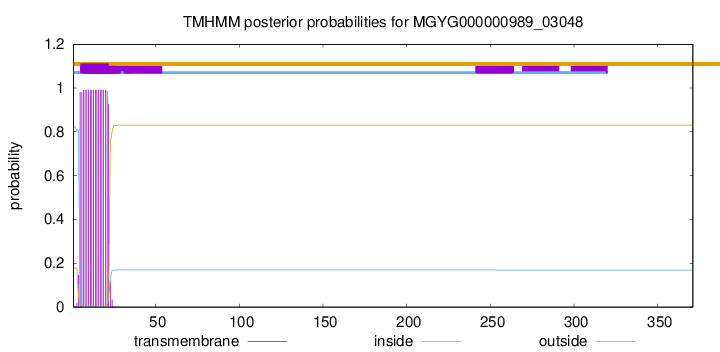
TMHMM annotations
Basic Information help
| Species | Niameybacter sp900549765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Cellulosilyticaceae; Niameybacter; Niameybacter sp900549765 | |||||||||||
| CAZyme ID | MGYG000000989_03048 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6548; End: 7663 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 48 | 370 | 9e-107 | 0.9906542056074766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 2.79e-09 | 77 | 176 | 27 | 130 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 2.68e-06 | 84 | 171 | 55 | 147 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK12845.1 | 6.39e-88 | 16 | 371 | 7 | 367 |
| QZY56115.1 | 1.56e-80 | 1 | 371 | 1 | 369 |
| QAA33952.1 | 9.01e-77 | 45 | 370 | 45 | 367 |
| ATD57594.1 | 6.16e-75 | 2 | 371 | 4 | 368 |
| SLK15954.1 | 6.16e-75 | 2 | 371 | 4 | 368 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 1.03e-64 | 36 | 371 | 15 | 345 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 6R2M_A | 1.51e-16 | 49 | 317 | 23 | 270 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 8.94e-20 | 48 | 370 | 44 | 351 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
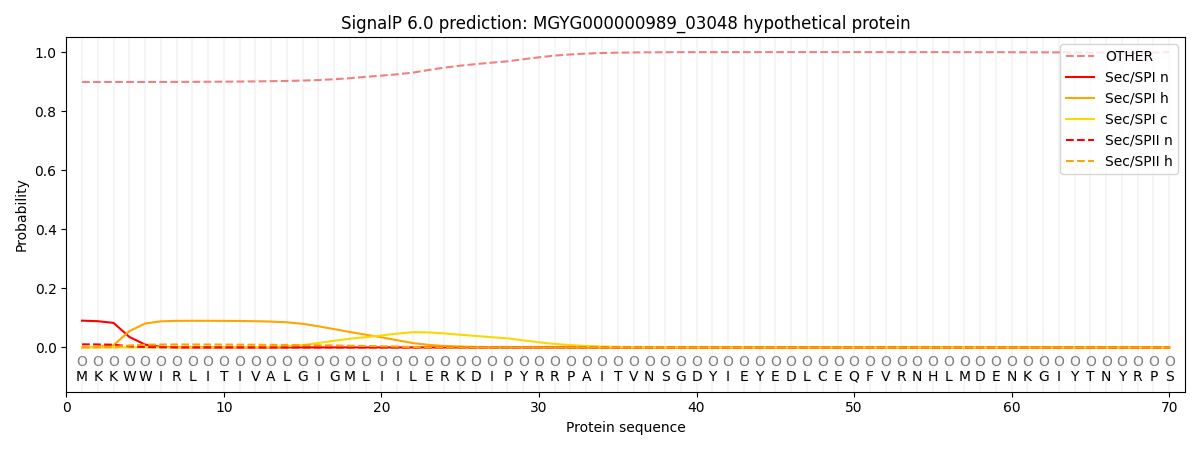
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.903388 | 0.083978 | 0.011205 | 0.000370 | 0.000258 | 0.000826 |