You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001004_01887
You are here: Home > Sequence: MGYG000001004_01887
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900539525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900539525 | |||||||||||
| CAZyme ID | MGYG000001004_01887 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2491; End: 5883 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 351 | 739 | 5.7e-26 | 0.3904761904761905 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 2.00e-19 | 91 | 240 | 4 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| COG3534 | AbfA | 5.17e-16 | 366 | 494 | 58 | 179 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 1.84e-11 | 794 | 919 | 67 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 5.42e-09 | 778 | 919 | 55 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| TIGR02243 | TIGR02243 | 1.56e-07 | 511 | 613 | 334 | 440 | putative baseplate assembly protein. This family consists of a large, conserved hypothetical protein in phage tail-like regions of at least six bacterial genomes: Gloeobacter violaceus PCC 7421, Geobacter sulfurreducens PCA, Streptomyces coelicolor A3(2), Streptomyces avermitilis MA-4680, Mesorhizobium loti, and Myxococcus xanthus. The C-terminal region is identified by the broader model pfam04865 as related to baseplate protein J from phage P2, but that relationship is not observed directly. [Mobile and extrachromosomal element functions, Prophage functions] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZO76045.1 | 1.67e-144 | 3 | 925 | 2 | 945 |
| AXM90856.1 | 6.00e-142 | 41 | 925 | 51 | 989 |
| BBA56144.1 | 8.06e-142 | 41 | 925 | 34 | 972 |
| BBA48000.1 | 1.56e-141 | 41 | 925 | 34 | 972 |
| ALE10461.1 | 1.56e-141 | 41 | 925 | 34 | 972 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VRK_A | 8.77e-07 | 366 | 494 | 60 | 179 | Structureof a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus],2VRK_B Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus],2VRK_C Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus [Thermobacillus xylanilyticus] |
| 2VRQ_A | 8.77e-07 | 366 | 494 | 60 | 179 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 6ZTA_A | 8.77e-07 | 366 | 496 | 60 | 183 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZT6_A | 1.52e-06 | 366 | 494 | 60 | 179 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZT8_A | 1.52e-06 | 366 | 494 | 60 | 179 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E7CY70 | 8.01e-11 | 366 | 509 | 70 | 202 | Exo-alpha-(1->6)-L-arabinofuranosidase OS=Bifidobacterium longum OX=216816 GN=afuB PE=1 SV=1 |
| P94531 | 1.47e-08 | 353 | 493 | 45 | 178 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase 1 OS=Bacillus subtilis (strain 168) OX=224308 GN=abfA PE=1 SV=2 |
| P53627 | 2.83e-07 | 366 | 493 | 58 | 180 | Intracellular exo-alpha-(1->5)-L-arabinofuranosidase OS=Streptomyces lividans OX=1916 GN=abfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
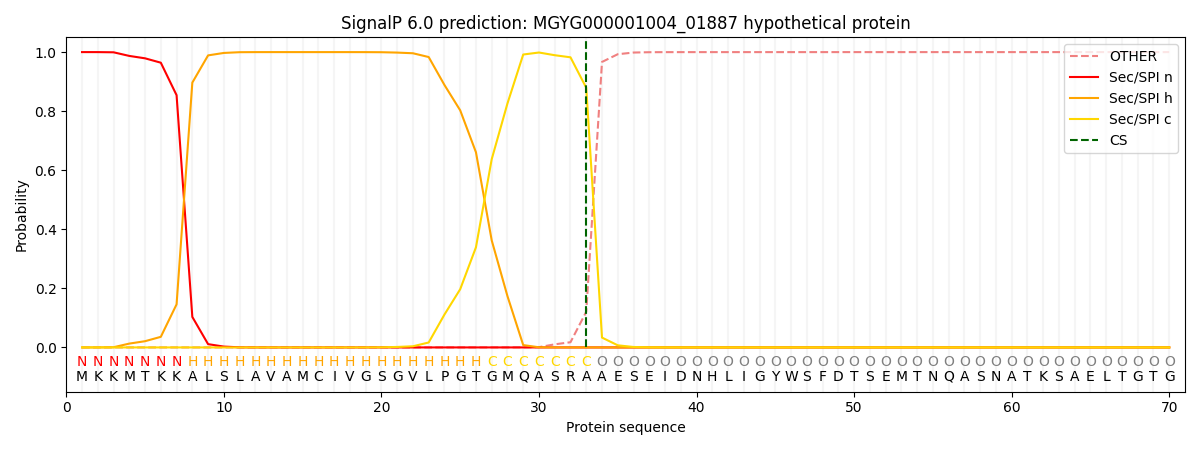
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000273 | 0.999010 | 0.000184 | 0.000200 | 0.000164 | 0.000142 |
