You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001005_01730
You are here: Home > Sequence: MGYG000001005_01730
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Faecalimonas sp900550235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas sp900550235 | |||||||||||
| CAZyme ID | MGYG000001005_01730 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73; End: 3084 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 321 | 434 | 1.5e-19 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 1.13e-22 | 869 | 1003 | 25 | 160 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG4193 | LytD | 1.07e-20 | 265 | 443 | 50 | 236 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5492 | YjdB | 3.55e-18 | 571 | 925 | 24 | 324 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam07538 | ChW | 6.94e-16 | 968 | 1002 | 1 | 35 | Clostridial hydrophobic W. A novel extracellular macromolecular system has been proposed based on the proteins containing ChW repeats. ChW stands for Clostridial hydrophobic with conserved W (tryptophan). This repeat was originally described in Clostridium acetobutylicum but is also found in other Gram-positive bacteria including Enterococcus faecalis, Streptococcus agalactiae and Streptomyces coelicolor. |
| smart00728 | ChW | 5.82e-15 | 967 | 1002 | 1 | 36 | Clostridial hydrophobic, with a conserved W residue, domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOV20091.1 | 7.59e-145 | 82 | 540 | 24 | 480 |
| QEK17141.1 | 7.29e-142 | 81 | 540 | 34 | 495 |
| AWY97555.1 | 5.78e-131 | 86 | 572 | 34 | 520 |
| QPS13527.1 | 1.30e-112 | 83 | 540 | 48 | 516 |
| QMW73215.1 | 1.30e-112 | 83 | 540 | 48 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.55e-31 | 171 | 443 | 55 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 2.10e-29 | 171 | 443 | 423 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.64e-29 | 171 | 443 | 467 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
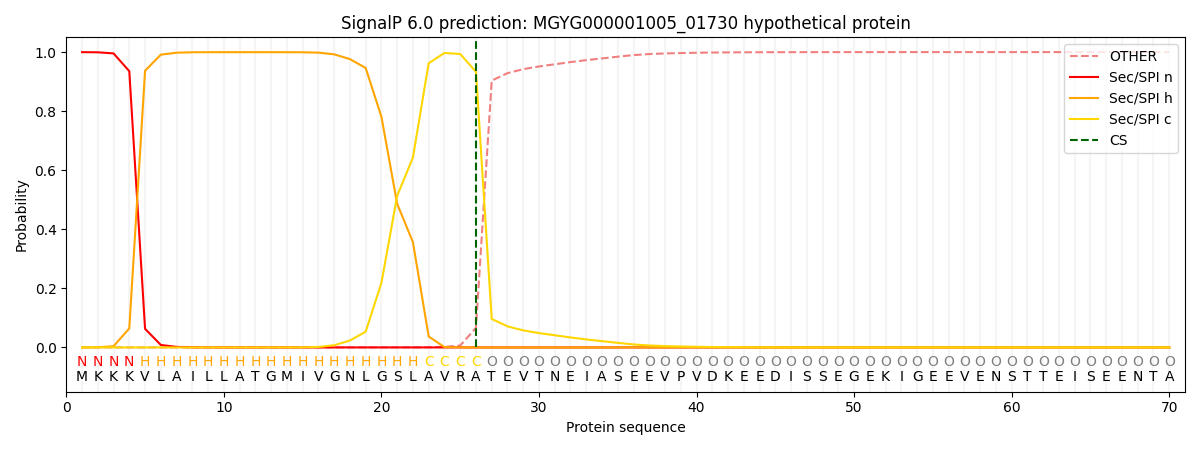
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001158 | 0.997814 | 0.000421 | 0.000204 | 0.000200 | 0.000179 |

