You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001007_01936
You are here: Home > Sequence: MGYG000001007_01936
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Intestinibacter sp900540355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Intestinibacter; Intestinibacter sp900540355 | |||||||||||
| CAZyme ID | MGYG000001007_01936 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5438; End: 7105 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 404 | 536 | 2.8e-23 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.49e-48 | 342 | 555 | 46 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG3103 | YgiM | 3.36e-22 | 1 | 156 | 1 | 158 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| COG3103 | YgiM | 3.66e-18 | 94 | 227 | 28 | 173 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| smart00047 | LYZ2 | 1.71e-16 | 402 | 534 | 11 | 136 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG3103 | YgiM | 1.10e-14 | 161 | 299 | 28 | 165 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYE98702.1 | 8.69e-147 | 28 | 555 | 261 | 811 |
| CEK37256.1 | 9.99e-145 | 28 | 555 | 342 | 899 |
| CEJ72783.1 | 9.91e-144 | 21 | 555 | 331 | 895 |
| AXU34918.1 | 5.59e-143 | 1 | 555 | 1 | 603 |
| AXU31130.1 | 5.59e-143 | 1 | 555 | 1 | 603 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WQW_A | 6.20e-40 | 320 | 541 | 23 | 257 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
| 6FXO_A | 2.97e-32 | 344 | 555 | 37 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 2.95e-29 | 381 | 535 | 65 | 209 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.96e-28 | 381 | 535 | 65 | 209 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 1.78e-23 | 341 | 534 | 73 | 256 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.39e-34 | 347 | 555 | 682 | 880 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q5HQB9 | 4.63e-31 | 347 | 555 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| O33635 | 4.63e-31 | 347 | 555 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q8CPQ1 | 8.26e-31 | 347 | 555 | 1131 | 1335 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q99V41 | 8.14e-29 | 344 | 555 | 1041 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
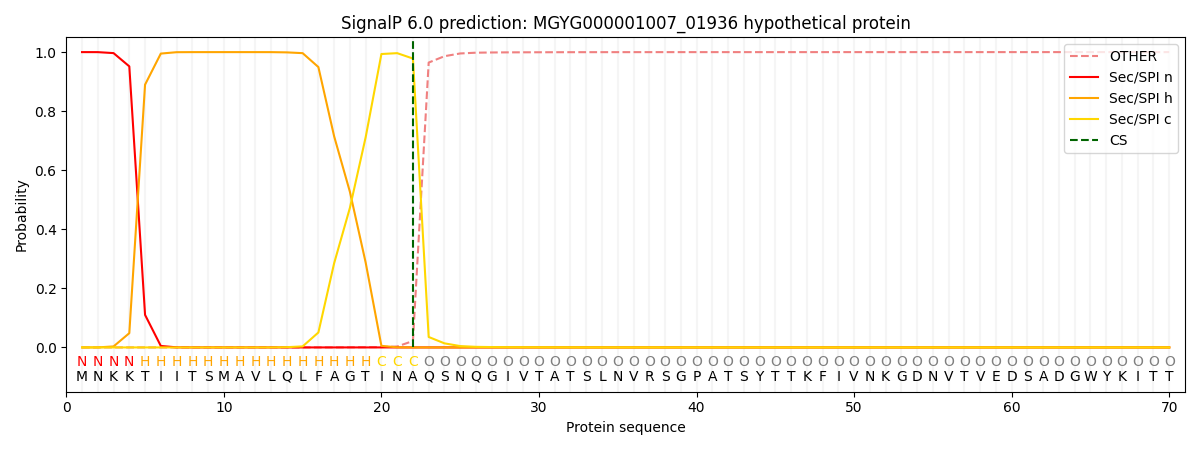
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000317 | 0.998837 | 0.000210 | 0.000210 | 0.000201 | 0.000166 |
