You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001008_01122
You are here: Home > Sequence: MGYG000001008_01122
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
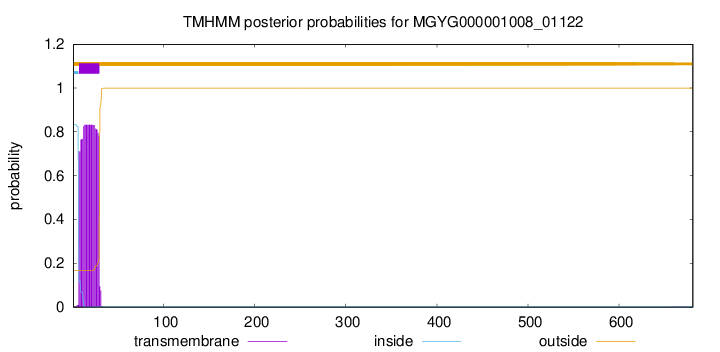
TMHMM annotations
Basic Information help
| Species | Clostridium_AQ sp900552125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Clostridium_AQ; Clostridium_AQ sp900552125 | |||||||||||
| CAZyme ID | MGYG000001008_01122 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52825; End: 54870 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 336 | 465 | 1.3e-20 | 0.9193548387096774 |
| CBM32 | 51 | 171 | 2.6e-20 | 0.8709677419354839 |
| CBM32 | 192 | 301 | 1.6e-19 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02838 | Glyco_hydro_20b | 7.10e-25 | 569 | 679 | 1 | 109 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 9.04e-15 | 48 | 170 | 6 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.29e-13 | 195 | 307 | 4 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 3.38e-11 | 339 | 445 | 6 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.55e-07 | 326 | 469 | 5 | 143 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58378.1 | 7.86e-176 | 186 | 680 | 34 | 531 |
| QWT17580.1 | 1.10e-92 | 187 | 679 | 40 | 564 |
| AMN35891.1 | 6.65e-76 | 166 | 680 | 21 | 542 |
| ALG48928.1 | 7.54e-75 | 166 | 680 | 21 | 544 |
| AQW23940.1 | 1.02e-74 | 166 | 680 | 21 | 544 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6JQF_A | 2.48e-19 | 479 | 680 | 3 | 194 | Crystallizationanalysis of a beta-N-acetylhexosaminidase (Am2136) from Akkermansia muciniphila [Akkermansia muciniphila ATCC BAA-835] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UPR7 | 6.07e-19 | 470 | 680 | 19 | 216 | Beta-hexosaminidase Amuc_2136 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_2136 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
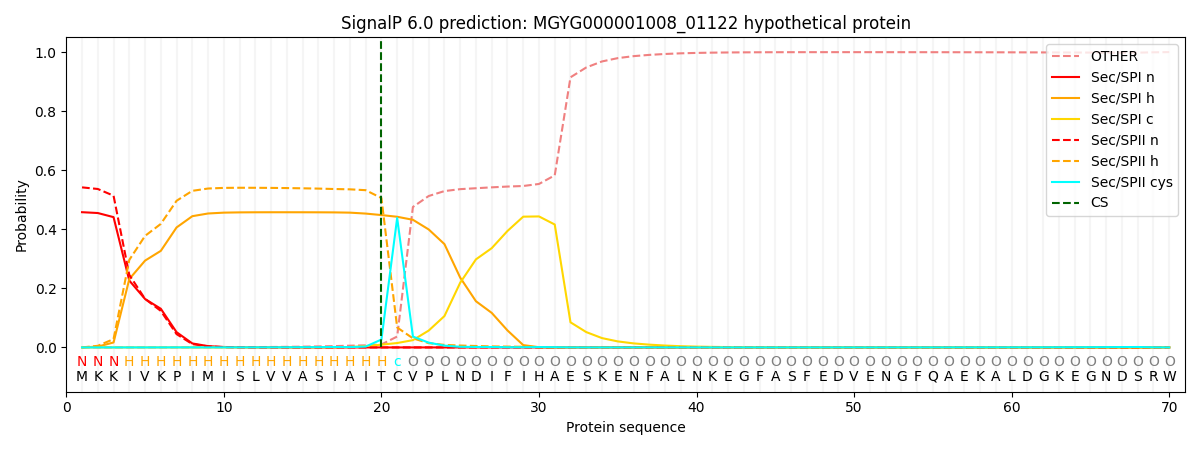
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000667 | 0.450451 | 0.548261 | 0.000228 | 0.000201 | 0.000174 |