You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001014_01097
You are here: Home > Sequence: MGYG000001014_01097
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
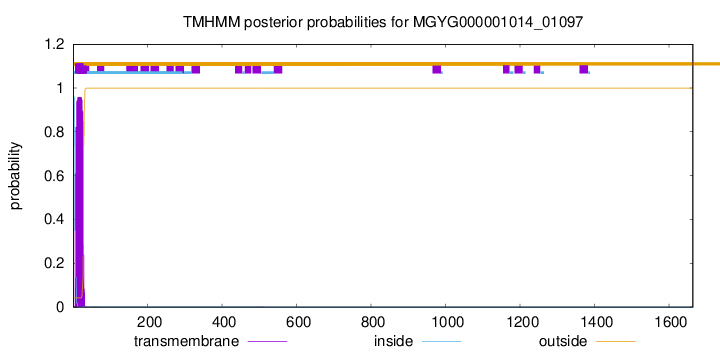
TMHMM annotations
Basic Information help
| Species | Varibaculum timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Varibaculum; Varibaculum timonense | |||||||||||
| CAZyme ID | MGYG000001014_01097 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 178; End: 5172 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 925 | 1056 | 1.7e-23 | 0.9516129032258065 |
| CBM32 | 780 | 904 | 3.7e-21 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 4.30e-51 | 1471 | 1662 | 1 | 163 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| pfam00754 | F5_F8_type_C | 1.76e-21 | 929 | 1056 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 7.29e-17 | 771 | 904 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.70e-14 | 938 | 1047 | 22 | 134 | Substituted updates: Jan 31, 2002 |
| cd00057 | FA58C | 1.06e-10 | 768 | 866 | 10 | 100 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH70491.1 | 1.07e-226 | 8 | 1144 | 5 | 1119 |
| SCG40654.1 | 2.29e-210 | 47 | 909 | 58 | 894 |
| AXI80327.1 | 1.03e-175 | 17 | 746 | 8 | 721 |
| QSY56848.1 | 2.20e-147 | 1160 | 1663 | 52 | 556 |
| QTB91571.1 | 1.63e-146 | 1160 | 1663 | 52 | 556 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EAE_A | 4.08e-74 | 1463 | 1664 | 11 | 213 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAD_A | 4.14e-74 | 1463 | 1664 | 12 | 214 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAB_A | 4.14e-74 | 1463 | 1664 | 12 | 214 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 4A41_A | 5.51e-17 | 765 | 907 | 25 | 160 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
| 2RDY_A | 2.79e-16 | 1502 | 1656 | 18 | 162 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 2.66e-17 | 1489 | 1664 | 55 | 221 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| I1S2N3 | 1.52e-14 | 760 | 907 | 45 | 187 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 2.00e-14 | 760 | 907 | 45 | 187 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| A2R797 | 2.37e-12 | 1502 | 1650 | 39 | 181 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q2USL3 | 1.79e-08 | 1504 | 1648 | 35 | 173 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
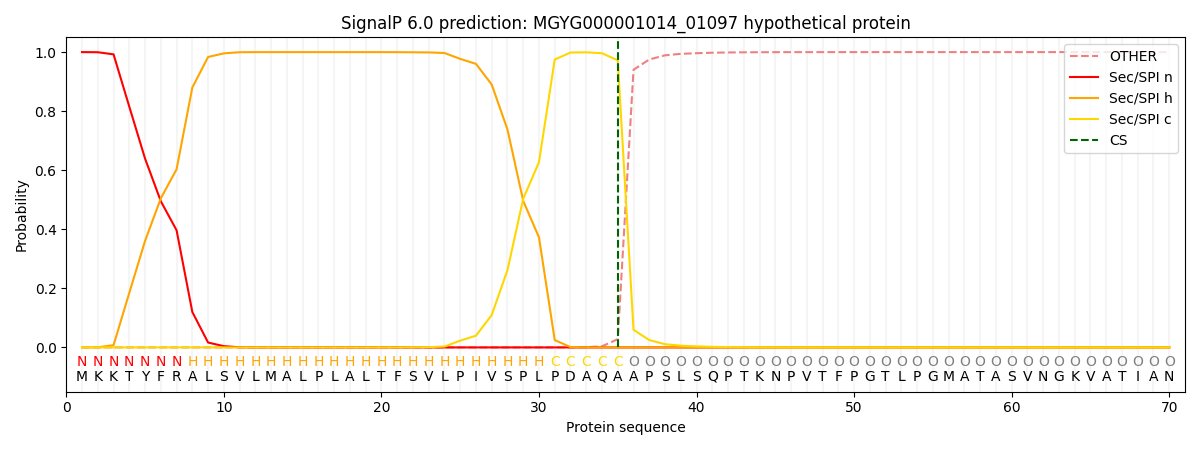
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.999009 | 0.000167 | 0.000199 | 0.000162 | 0.000145 |