You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001016_01917
You are here: Home > Sequence: MGYG000001016_01917
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
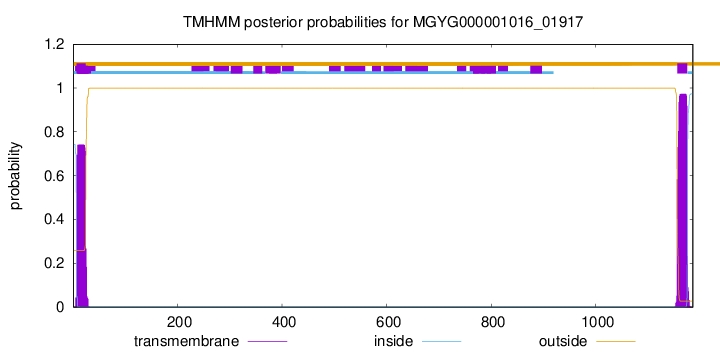
TMHMM annotations
Basic Information help
| Species | UMGS1537 sp900543695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; UBA1212; UBA1255; UMGS1537; UMGS1537 sp900543695 | |||||||||||
| CAZyme ID | MGYG000001016_01917 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26074; End: 29634 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 686 | 944 | 1.4e-23 | 0.6430769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 6.38e-23 | 630 | 926 | 77 | 367 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam06439 | DUF1080 | 0.003 | 248 | 355 | 58 | 177 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 1.01e-172 | 373 | 1057 | 71 | 745 |
| QCD41820.1 | 9.75e-108 | 336 | 994 | 180 | 834 |
| ADB48561.1 | 1.92e-42 | 626 | 1089 | 25 | 466 |
| QGQ94767.1 | 4.50e-38 | 626 | 1106 | 34 | 508 |
| QYH35737.1 | 8.77e-38 | 625 | 1126 | 122 | 631 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MXN_A | 2.05e-09 | 634 | 800 | 25 | 166 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 5OLP_A | 1.02e-08 | 622 | 922 | 38 | 324 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
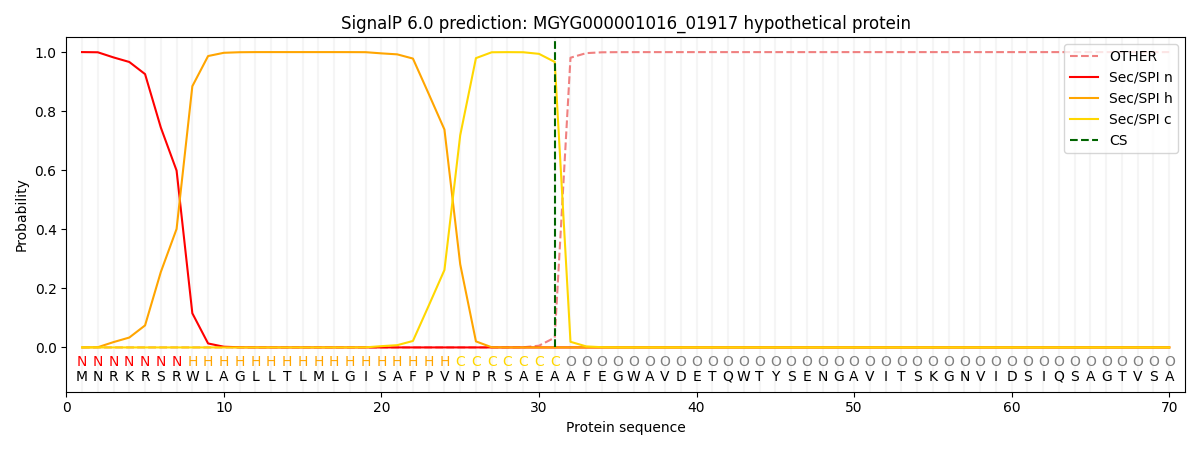
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000224 | 0.999113 | 0.000173 | 0.000186 | 0.000157 | 0.000137 |