You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001018_00230
You are here: Home > Sequence: MGYG000001018_00230
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
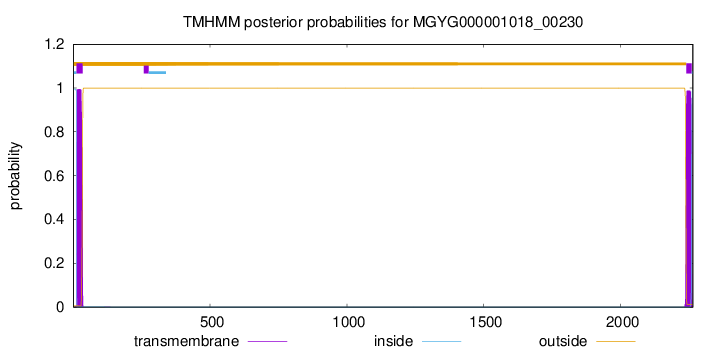
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900751785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900751785 | |||||||||||
| CAZyme ID | MGYG000001018_00230 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14220; End: 21014 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1061 | 1181 | 2.3e-17 | 0.8656716417910447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.57e-19 | 1621 | 1766 | 7 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.96e-15 | 1057 | 1185 | 4 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.83e-14 | 1634 | 1766 | 35 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam18998 | Flg_new_2 | 4.35e-13 | 1868 | 1935 | 1 | 71 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| smart00776 | NPCBM | 8.99e-13 | 1058 | 1185 | 10 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYJ80995.1 | 2.05e-142 | 30 | 1184 | 16 | 1064 |
| QYJ92352.1 | 1.78e-141 | 30 | 1184 | 16 | 1064 |
| QUH30699.1 | 2.42e-139 | 67 | 1399 | 37 | 1009 |
| ABO24155.1 | 1.15e-136 | 30 | 1184 | 16 | 1064 |
| AQM60758.1 | 3.43e-52 | 1461 | 2150 | 1360 | 1999 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NIT_A | 1.68e-06 | 1553 | 1605 | 902 | 954 | ChainA, Beta-galactosidase [Bifidobacterium bifidum],7NIT_B Chain B, Beta-galactosidase [Bifidobacterium bifidum],7NIT_C Chain C, Beta-galactosidase [Bifidobacterium bifidum],7NIT_D Chain D, Beta-galactosidase [Bifidobacterium bifidum],7NIT_E Chain E, Beta-galactosidase [Bifidobacterium bifidum],7NIT_F Chain F, Beta-galactosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.84e-11 | 1462 | 2067 | 1298 | 1868 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
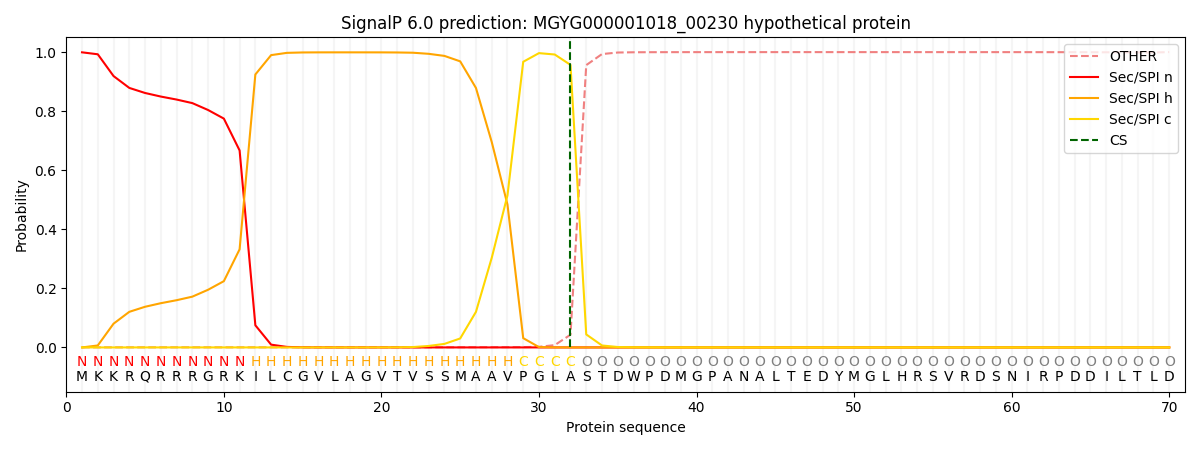
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002109 | 0.995971 | 0.000556 | 0.000771 | 0.000313 | 0.000240 |