You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001021_01606
You are here: Home > Sequence: MGYG000001021_01606
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp000980705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp000980705 | |||||||||||
| CAZyme ID | MGYG000001021_01606 | |||||||||||
| CAZy Family | CBM30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15150; End: 18659 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 301 | 772 | 7.4e-70 | 0.9952153110047847 |
| PL31 | 858 | 1049 | 5.4e-65 | 0.9946524064171123 |
| CBM30 | 57 | 201 | 8.6e-17 | 0.8352941176470589 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 6.64e-60 | 303 | 768 | 1 | 371 | Glycosyl hydrolase family 9. |
| pfam02927 | CelD_N | 1.57e-16 | 184 | 269 | 1 | 82 | Cellulase N-terminal ig-like domain. |
| cd14251 | PL-6 | 8.19e-16 | 851 | 1108 | 1 | 270 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| cd02850 | E_set_Cellulase_N | 1.67e-14 | 186 | 296 | 2 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| cd14256 | Dockerin_I | 3.03e-14 | 785 | 839 | 2 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL51370.1 | 7.21e-99 | 68 | 806 | 101 | 787 |
| AEV69564.1 | 2.02e-97 | 53 | 808 | 87 | 786 |
| AVQ18252.1 | 1.78e-89 | 60 | 774 | 102 | 773 |
| ABN51859.1 | 9.54e-88 | 68 | 782 | 103 | 759 |
| ADU74665.1 | 9.54e-88 | 68 | 782 | 103 | 759 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KFN_A | 2.36e-75 | 857 | 1120 | 14 | 275 | Crystalstructure of alginate lyase from Paenibacillus sp. str. FPU-7 [Paenibacillus sp. FPU-7] |
| 1UT9_A | 1.48e-37 | 184 | 779 | 5 | 609 | ChainA, CELLULOSE 1,4-BETA-CELLOBIOSIDASE [Acetivibrio thermocellus] |
| 1RQ5_A | 1.14e-36 | 184 | 777 | 5 | 607 | ChainA, Cellobiohydrolase [Acetivibrio thermocellus] |
| 5U0H_A | 3.55e-34 | 210 | 771 | 13 | 537 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 5U2O_A | 2.00e-33 | 210 | 771 | 13 | 537 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23658 | 5.39e-45 | 186 | 775 | 4 | 546 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| A3DCH1 | 2.23e-40 | 184 | 806 | 212 | 854 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
| P0C2S1 | 2.23e-40 | 184 | 806 | 212 | 854 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus OX=1515 GN=celK PE=1 SV=1 |
| A3DDN1 | 5.01e-32 | 159 | 806 | 28 | 605 | Endoglucanase D OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celD PE=1 SV=1 |
| P0C2S4 | 5.51e-32 | 159 | 806 | 4 | 581 | Endoglucanase D (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
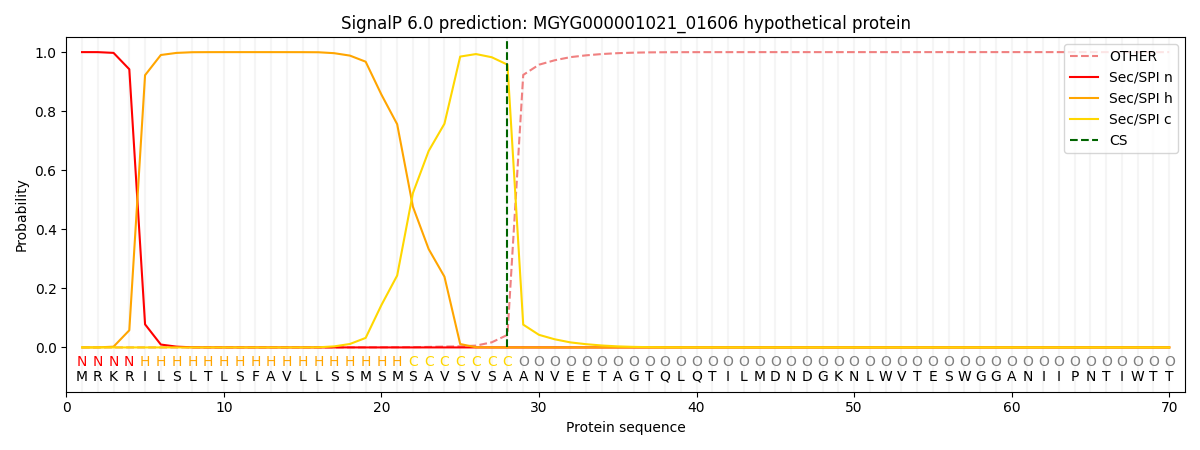
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000346 | 0.998804 | 0.000182 | 0.000274 | 0.000187 | 0.000155 |
