You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001021_01618
You are here: Home > Sequence: MGYG000001021_01618
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
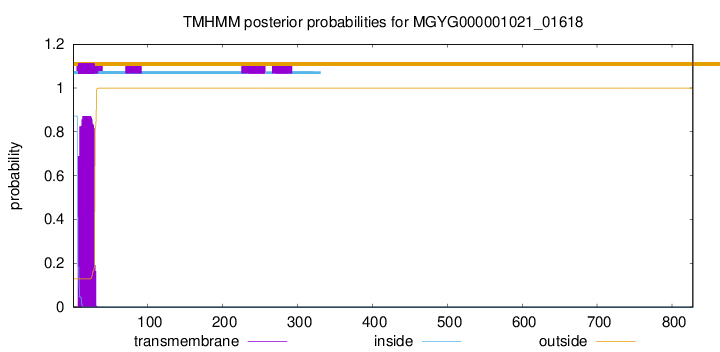
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp000980705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp000980705 | |||||||||||
| CAZyme ID | MGYG000001021_01618 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34120; End: 36606 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 96 | 377 | 7.3e-81 | 0.9930795847750865 |
| CBM23 | 506 | 669 | 1e-44 | 0.9938271604938271 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 8.18e-22 | 88 | 392 | 14 | 289 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 2.29e-15 | 501 | 669 | 1 | 173 | Carbohydrate binding domain (family 11). |
| cd14256 | Dockerin_I | 7.68e-15 | 762 | 818 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 4.29e-09 | 763 | 817 | 1 | 55 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam00150 | Cellulase | 5.16e-07 | 102 | 357 | 30 | 252 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL52789.1 | 4.37e-205 | 38 | 676 | 610 | 1340 |
| BAV13033.1 | 4.37e-205 | 38 | 676 | 610 | 1340 |
| ADZ85047.1 | 4.57e-191 | 40 | 552 | 630 | 1157 |
| QEH70547.1 | 4.30e-187 | 40 | 552 | 630 | 1157 |
| AEY66038.1 | 1.17e-143 | 40 | 409 | 25 | 394 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RH9_A | 2.28e-52 | 48 | 385 | 6 | 344 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
| 4QP0_A | 2.95e-43 | 48 | 376 | 4 | 325 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3PZ9_A | 1.12e-39 | 53 | 375 | 18 | 340 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 9.67e-38 | 53 | 375 | 1 | 326 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 1QNO_A | 1.05e-31 | 48 | 399 | 4 | 329 | ChainA, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6Z310 | 3.46e-58 | 38 | 407 | 28 | 397 | Putative mannan endo-1,4-beta-mannosidase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN9 PE=2 SV=2 |
| Q0JKM9 | 1.01e-54 | 47 | 376 | 36 | 372 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
| Q9FJZ3 | 1.35e-54 | 41 | 385 | 23 | 372 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
| Q9SG94 | 4.39e-54 | 42 | 385 | 25 | 375 | Mannan endo-1,4-beta-mannosidase 3 OS=Arabidopsis thaliana OX=3702 GN=MAN3 PE=2 SV=1 |
| Q9FZ29 | 7.71e-54 | 43 | 419 | 24 | 401 | Mannan endo-1,4-beta-mannosidase 1 OS=Arabidopsis thaliana OX=3702 GN=MAN1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
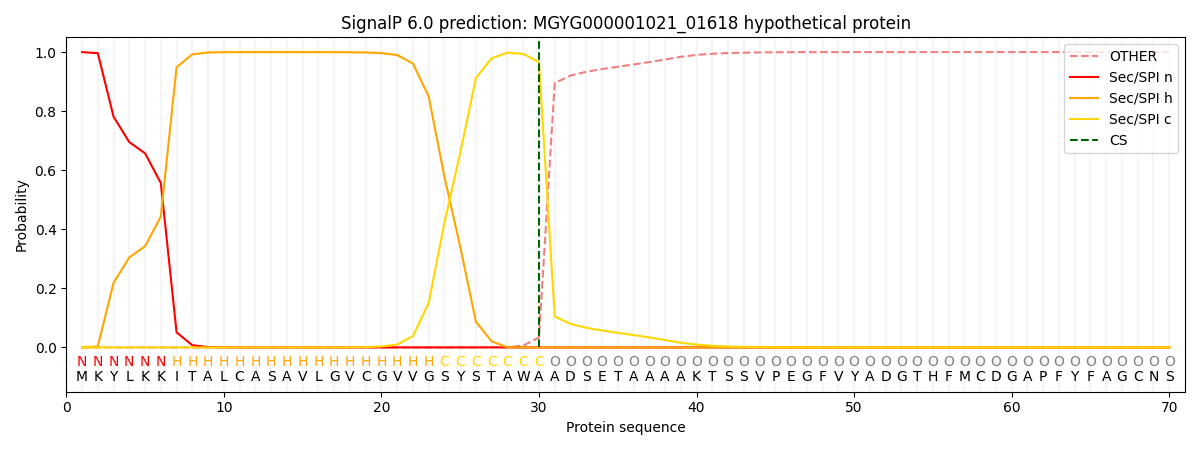
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000407 | 0.998553 | 0.000445 | 0.000232 | 0.000177 | 0.000148 |