You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001024_01265
You are here: Home > Sequence: MGYG000001024_01265
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
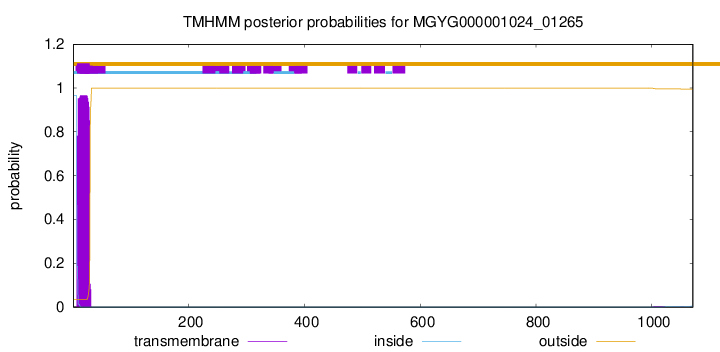
TMHMM annotations
Basic Information help
| Species | HGM11521 sp900751885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; HGM11521; HGM11521 sp900751885 | |||||||||||
| CAZyme ID | MGYG000001024_01265 | |||||||||||
| CAZy Family | CE6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43436; End: 46651 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE6 | 107 | 205 | 1.1e-23 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03629 | SASA | 1.80e-59 | 33 | 281 | 2 | 226 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| pfam12733 | Cadherin-like | 1.51e-06 | 793 | 867 | 9 | 86 | Cadherin-like beta sandwich domain. This domain is found in several bacterial, metazoan and chlorophyte algal proteins. A profile-profile comparison recovered the cadherin domain and a comparison of the predicted structure of this domain with the crystal structure of the cadherin showed a congruent seven stranded secondary structure. The domain is widespread in bacteria and seen in the firmicutes, actinobacteria, certain proteobacteria, bacteroides and chlamydiae with an expansion in Clostridium. In contrast, it is limited in its distribution in eukaryotes suggesting that it was derived through lateral transfer from bacteria. In prokaryotes, this domain is widely fused to other domains such as FNIII (Fibronectin Type III), TIG, SLH (S-layer homology), discoidin, cell-wall-binding repeat domain and alpha-amylase-like glycohydrolases. These associations are suggestive of a carbohydrate-binding function for this cadherin-like domain. In animal proteins it is associated with an ATP-grasp domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT42077.1 | 4.29e-48 | 34 | 285 | 32 | 267 |
| BCA48725.1 | 1.08e-47 | 34 | 285 | 32 | 267 |
| QMW86523.1 | 1.48e-47 | 34 | 285 | 32 | 267 |
| QUT69878.1 | 1.48e-47 | 34 | 285 | 32 | 267 |
| AAO79285.1 | 1.48e-47 | 34 | 285 | 32 | 267 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2APJ_A | 1.45e-14 | 105 | 272 | 91 | 248 | X-RayStructure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_B X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_C X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana],2APJ_D X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L9J9 | 1.77e-14 | 35 | 272 | 24 | 248 | Probable carbohydrate esterase At4g34215 OS=Arabidopsis thaliana OX=3702 GN=At4g34215 PE=1 SV=2 |
| D5EXZ4 | 3.73e-10 | 30 | 294 | 41 | 306 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
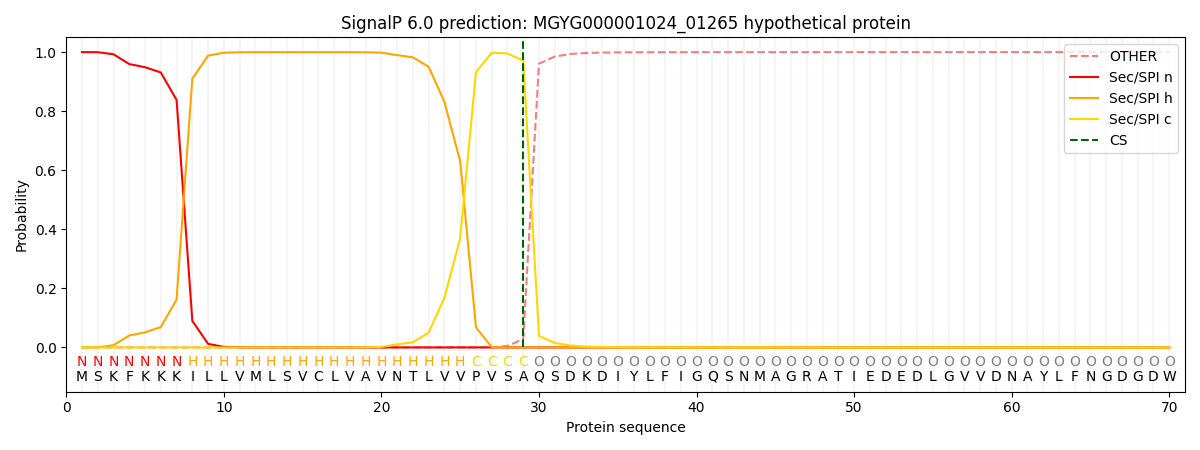
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000437 | 0.998774 | 0.000274 | 0.000171 | 0.000156 | 0.000154 |