You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001024_01635
You are here: Home > Sequence: MGYG000001024_01635
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM11521 sp900751885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; HGM11521; HGM11521 sp900751885 | |||||||||||
| CAZyme ID | MGYG000001024_01635 | |||||||||||
| CAZy Family | CE12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67541; End: 73867 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 755 | 1440 | 1.8e-54 | 0.6914893617021277 |
| CE12 | 415 | 619 | 1.3e-31 | 0.9904761904761905 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01821 | Rhamnogalacturan_acetylesterase_like | 5.37e-31 | 415 | 620 | 2 | 198 | Rhamnogalacturan_acetylesterase_like subgroup of SGNH-hydrolases. Rhamnogalacturan acetylesterase removes acetyl esters from rhamnogalacturonan substrates, and renders them susceptible to degradation by rhamnogalacturonases. Rhamnogalacturonans are highly branched regions in pectic polysaccharides, consisting of repeating -(1,2)-L-Rha-(1,4)-D-GalUA disaccharide units, with many rhamnose residues substituted by neutral oligosaccharides such as arabinans, galactans and arabinogalactans. Extracellular enzymes participating in the degradation of plant cell wall polymers, such as Rhamnogalacturonan acetylesterase, would typically be found in saprophytic and plant pathogenic fungi and bacteria. |
| COG3250 | LacZ | 1.13e-17 | 759 | 1184 | 9 | 428 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| cd04084 | CBM6_xylanase-like | 7.36e-16 | 1703 | 1813 | 15 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| PRK10150 | PRK10150 | 4.56e-15 | 755 | 1183 | 4 | 443 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 7.00e-13 | 925 | 1037 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBE17052.1 | 4.30e-183 | 784 | 1677 | 58 | 948 |
| QGY48213.1 | 1.15e-180 | 761 | 1673 | 29 | 946 |
| QCY56657.1 | 3.77e-167 | 761 | 1691 | 24 | 938 |
| QUT34134.1 | 6.98e-167 | 761 | 1677 | 27 | 923 |
| ABR43180.1 | 7.18e-167 | 761 | 1691 | 24 | 938 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KGY_A | 3.28e-10 | 754 | 1136 | 9 | 374 | ChainA, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_B Chain B, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_C Chain C, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_D Chain D, Beta-glucuronidase [Faecalibacterium prausnitzii] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
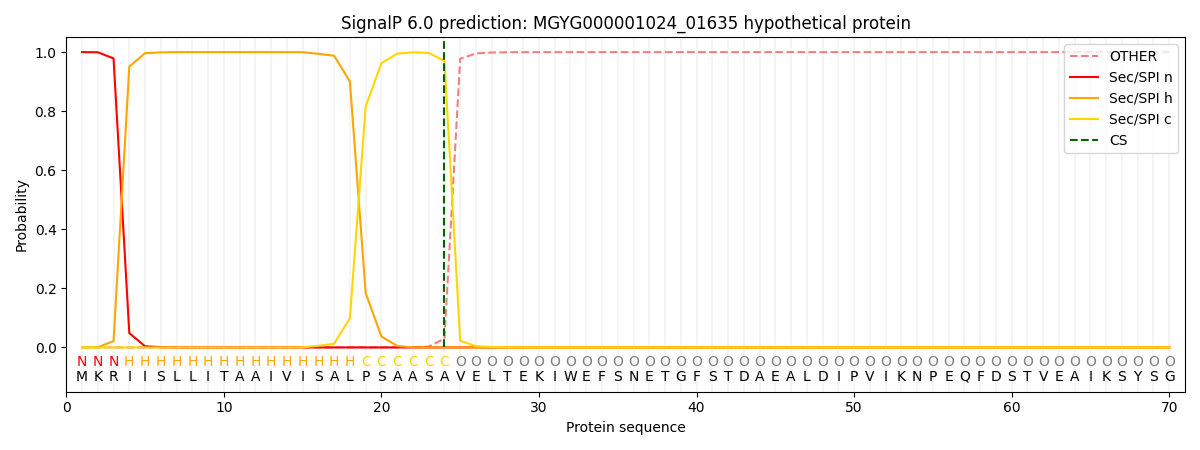
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000194 | 0.999135 | 0.000156 | 0.000186 | 0.000157 | 0.000145 |
