You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001027_01370
You are here: Home > Sequence: MGYG000001027_01370
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
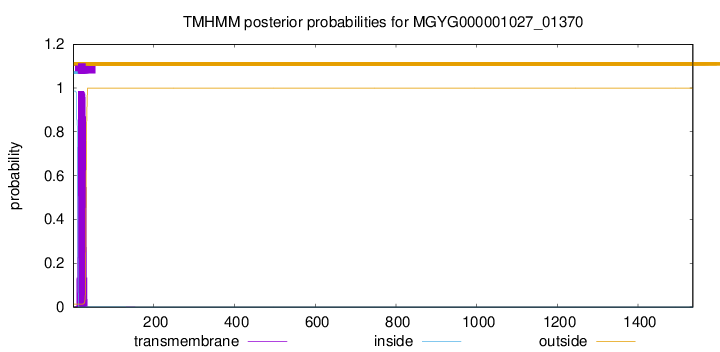
TMHMM annotations
Basic Information help
| Species | CAG-56 sp900752065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-56; CAG-56 sp900752065 | |||||||||||
| CAZyme ID | MGYG000001027_01370 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45101; End: 49711 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 182 | 481 | 5.2e-90 | 0.9627118644067797 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.48e-105 | 182 | 476 | 1 | 283 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 2.98e-23 | 38 | 175 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| sd00036 | LRR_3 | 3.95e-16 | 1444 | 1513 | 1 | 68 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 6.17e-15 | 1444 | 1515 | 47 | 116 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 1.88e-14 | 1447 | 1518 | 2 | 69 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQW22579.1 | 1.05e-302 | 16 | 1200 | 12 | 1351 |
| AXH51290.1 | 1.52e-302 | 16 | 1200 | 12 | 1351 |
| ATD49893.1 | 1.52e-302 | 16 | 1200 | 12 | 1351 |
| BAB79897.1 | 8.37e-302 | 16 | 1200 | 12 | 1351 |
| ASY50288.1 | 1.66e-301 | 16 | 1200 | 12 | 1351 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV4_A | 1.48e-183 | 39 | 656 | 32 | 653 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PWI_A | 7.06e-103 | 29 | 649 | 23 | 626 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV5_A | 1.27e-56 | 38 | 610 | 39 | 596 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 3.21e-50 | 43 | 786 | 18 | 718 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
| 2XPK_A | 6.01e-49 | 43 | 483 | 18 | 458 | Cell-penetrant,nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens],2XPK_B Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 1.67e-302 | 16 | 1200 | 12 | 1351 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q8XL08 | 2.81e-49 | 43 | 786 | 48 | 748 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 1.51e-48 | 43 | 786 | 48 | 748 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 2.45e-47 | 128 | 572 | 100 | 520 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q8VIJ5 | 2.83e-20 | 183 | 450 | 63 | 329 | Protein O-GlcNAcase OS=Rattus norvegicus OX=10116 GN=Oga PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
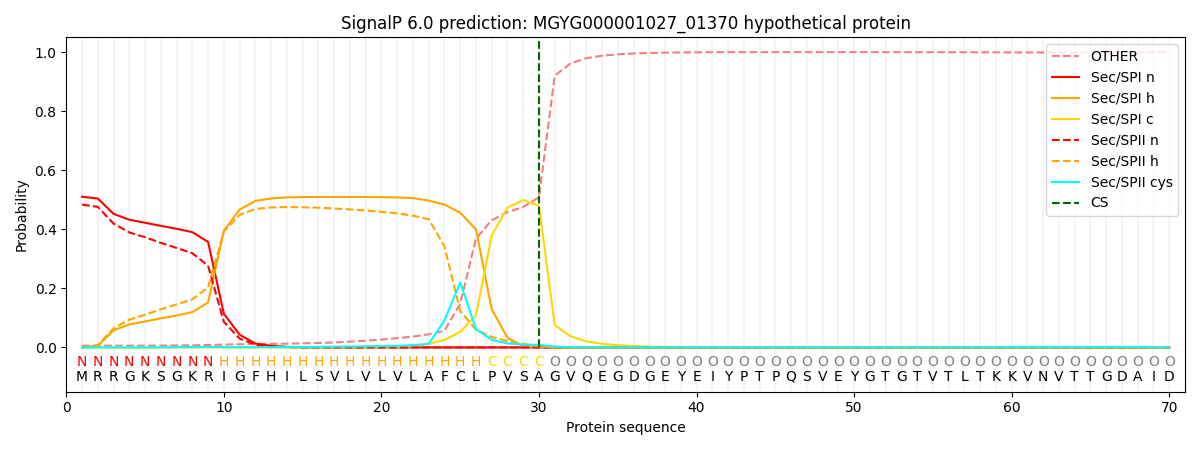
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006990 | 0.501880 | 0.490143 | 0.000498 | 0.000258 | 0.000226 |