You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001034_00504
You are here: Home > Sequence: MGYG000001034_00504
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900554695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900554695 | |||||||||||
| CAZyme ID | MGYG000001034_00504 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36717; End: 40679 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 176 | 505 | 9.1e-49 | 0.8687150837988827 |
| CBM6 | 552 | 674 | 5.9e-28 | 0.8623188405797102 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04084 | CBM6_xylanase-like | 2.13e-33 | 549 | 673 | 1 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| smart00606 | CBD_IV | 1.61e-29 | 545 | 673 | 2 | 129 | Cellulose Binding Domain Type IV. |
| pfam03663 | Glyco_hydro_76 | 2.45e-27 | 208 | 482 | 36 | 313 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| pfam03422 | CBM_6 | 1.23e-25 | 552 | 674 | 1 | 124 | Carbohydrate binding module (family 6). |
| COG4833 | COG4833 | 3.56e-12 | 208 | 466 | 46 | 281 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74831.1 | 5.94e-256 | 1 | 1320 | 1 | 1310 |
| QIA07414.1 | 3.31e-92 | 17 | 673 | 60 | 671 |
| QOV19890.1 | 3.58e-59 | 205 | 676 | 56 | 495 |
| AYD48939.1 | 6.63e-58 | 20 | 498 | 27 | 472 |
| QEC70572.1 | 1.11e-57 | 18 | 535 | 25 | 494 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M77_A | 1.54e-47 | 208 | 675 | 73 | 524 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
| 4BOK_A | 5.52e-32 | 208 | 506 | 39 | 331 | ChainA, Alpha-1,6-mannanase [Niallia circulans] |
| 4BOJ_A | 5.88e-32 | 208 | 506 | 42 | 334 | ChainA, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
| 4D4A_A | 2.84e-31 | 208 | 506 | 60 | 352 | ChainA, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
| 5AGD_A | 1.28e-30 | 208 | 506 | 60 | 352 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5AGD_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBM_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33558 | 9.14e-14 | 539 | 696 | 241 | 394 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| Q45071 | 3.70e-13 | 548 | 674 | 381 | 511 | Arabinoxylan arabinofuranohydrolase OS=Bacillus subtilis (strain 168) OX=224308 GN=xynD PE=1 SV=2 |
| Q8GJ44 | 2.62e-12 | 535 | 674 | 513 | 647 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P10478 | 1.62e-11 | 535 | 716 | 286 | 463 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
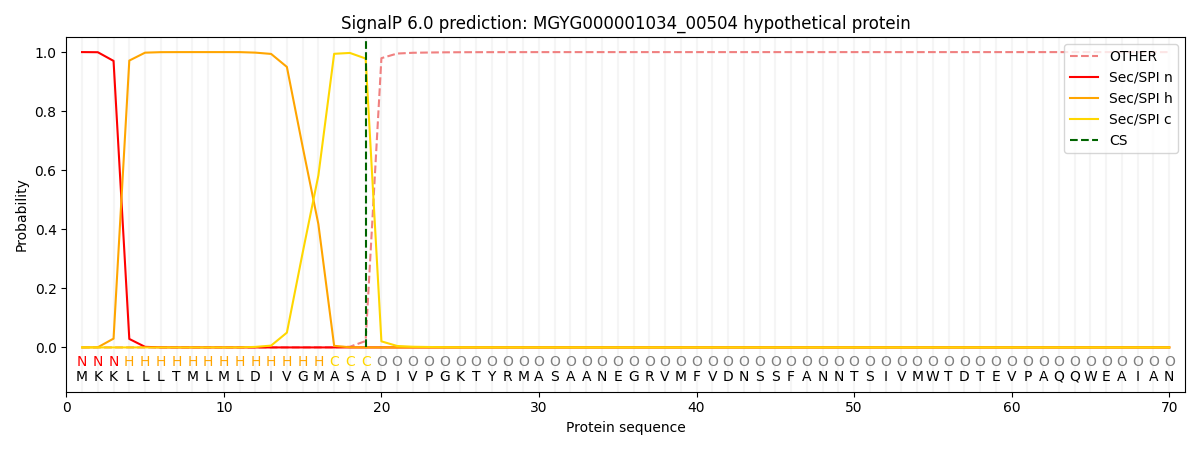
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000278 | 0.999063 | 0.000176 | 0.000178 | 0.000156 | 0.000143 |
