You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001034_01387
You are here: Home > Sequence: MGYG000001034_01387
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900554695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900554695 | |||||||||||
| CAZyme ID | MGYG000001034_01387 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5296; End: 8232 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 550 | 933 | 7.4e-42 | 0.6507936507936508 |
| CBM66 | 72 | 227 | 1.8e-29 | 0.9741935483870968 |
| CBM66 | 247 | 399 | 1.2e-23 | 0.9548387096774194 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 4.17e-18 | 562 | 934 | 62 | 501 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 2.47e-17 | 745 | 925 | 3 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06439 | DUF1080 | 3.28e-13 | 88 | 230 | 33 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| pfam06964 | Alpha-L-AF_C | 2.02e-11 | 816 | 925 | 67 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06439 | DUF1080 | 2.33e-11 | 254 | 402 | 25 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL02085.1 | 2.48e-201 | 18 | 914 | 25 | 789 |
| BBL12837.1 | 1.54e-199 | 18 | 914 | 25 | 789 |
| BBL10044.1 | 1.54e-199 | 18 | 914 | 25 | 789 |
| CUA16970.1 | 4.77e-188 | 16 | 929 | 15 | 795 |
| AKA54044.1 | 9.49e-188 | 16 | 929 | 15 | 795 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 2.41e-08 | 499 | 928 | 147 | 619 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SG80 | 5.56e-25 | 447 | 950 | 138 | 670 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 8.38e-23 | 449 | 942 | 139 | 659 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| U6A629 | 5.89e-20 | 441 | 692 | 97 | 391 | Alpha-L-arabinofuranosidase A OS=Penicillium canescens OX=5083 GN=abfA PE=1 SV=1 |
| Q0CTV2 | 2.44e-16 | 420 | 929 | 78 | 625 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=abfA PE=3 SV=1 |
| B8NKA3 | 7.37e-13 | 460 | 884 | 122 | 582 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
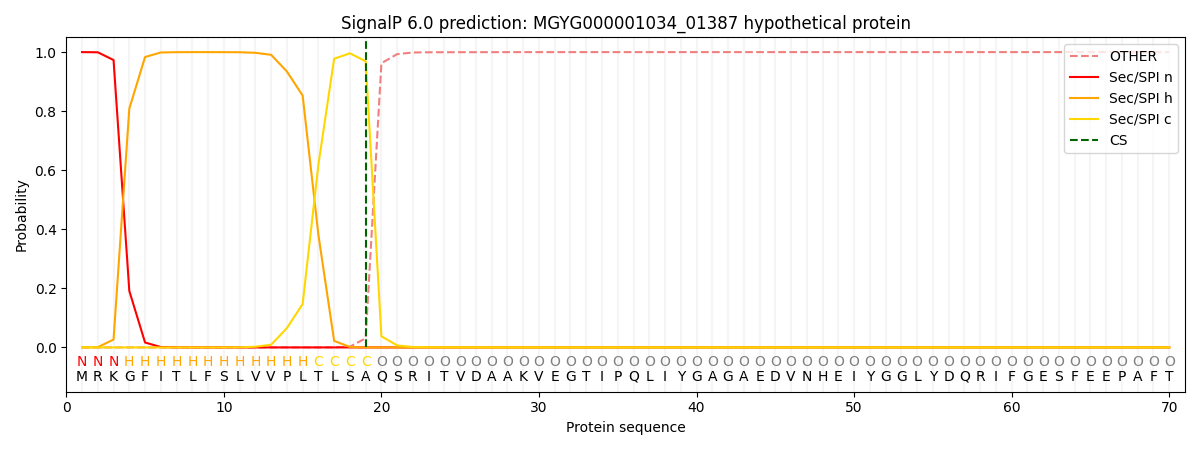
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000452 | 0.998589 | 0.000403 | 0.000192 | 0.000184 | 0.000160 |
