You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001039_00425
You are here: Home > Sequence: MGYG000001039_00425
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900553185 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900553185 | |||||||||||
| CAZyme ID | MGYG000001039_00425 | |||||||||||
| CAZy Family | GH97 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 183272; End: 185974 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH97 | 10 | 653 | 1.8e-191 | 0.9920760697305864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam10566 | Glyco_hydro_97 | 1.38e-133 | 290 | 555 | 1 | 278 | Glycoside hydrolase 97. This domain is the catalytic region of the bacterial glycosyl-hydrolase family 97. This central part of the GH97 family protein sequences represents a typical and complete (beta/alpha)8-barrel or catalytic TIM-barrel type domain. The N- and C-terminal parts of the sequences, mainly consisting of beta-strands, form two additional non-catalytic domains. In all known glycosidases with the (beta-alpha)8-barrel fold, the amino acid residues at the active site are located on the C-termini of the beta-strands. |
| pfam14508 | GH97_N | 2.08e-57 | 28 | 285 | 1 | 235 | Glycosyl-hydrolase 97 N-terminal. This N-terminal domain of glycosyl-hydrolase-97 contributes part of the active site pocket. It is also important for contact with the catalytic and C-terminal domains of the whole. |
| pfam14509 | GH97_C | 6.47e-42 | 559 | 653 | 2 | 96 | Glycosyl-hydrolase 97 C-terminal, oligomerization. Glycosyl-hydrolase-97 is made up of three tightly linked and highly conserved globular domains. The C-terminal domain is found to be necessary for oligomerization of the whole molecule in order to create the active-site pocket and the Ca++-binding site. |
| cd20169 | Peptidase_M90_mtfA | 1.53e-05 | 793 | 887 | 115 | 208 | Mlc titration factor A (MtfA) is a zinc metallopeptidase (M90 peptidase). This subfamily includes the Mlc Titration Factor A (MtfA; also known as YeeI or DgsA anti-repressor MtfA) which is involved in the control of the glucose-phosphotransferase sensory and regulatory system by inactivation of the repressor Mlc (making large colonies). It can cleave synthetic substrates of both carboxypeptidases and aminopeptidases, with strongest activity towards the latter. Its biologically relevant substrate has yet to be identified. Although it interacts with the transcription repressor Mlc, it does not cleave it. However, Mlc seems to activate the peptidase activity of MtfA. MtfA is related to the catalytic domain of the anthrax lethal factor which is a zinc-dependent metalloprotease, targeting mitogen-activated protein kinase kinases (MAPKKs), and resulting in apoptosis, as well as the Mop (modulation of pathogenesis) protein involved in the virulence of Vibrio cholerae; although sequence similarity is low, conservation is observed in the overall structure as well as in the residues around the active site. |
| pfam06167 | Peptidase_M90 | 6.38e-04 | 780 | 887 | 141 | 242 | Glucose-regulated metallo-peptidase M90. MtfA (earlier known as YeeI) is a transcription factor A that binds Mlc (make large colonies), itself a repressor of glucose and hence a protein important in regulation of the phosphoenolpyruvate:glucose-phosphotransferase (ptsG) system, the major glucose transporter in E.coli. Mlc is a repressor of ptsG, and MtfA is found to bind and inactivate Mlc with high affinity. The membrane-bound protein EIICBGlc encoded by the ptsG gene is the major glucose transporter in Escherichia coli. MtfA is found to be a glucose-regulated peptidase, whose activity is regulated by binding to Mlc available in the cytoplasm, which in turn has been released from EIICBGlc during times when no glucose is taken up. A physiologically relevant target for this peptidase is not yet known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJR66042.1 | 0.0 | 22 | 899 | 28 | 906 |
| QJR70731.1 | 0.0 | 22 | 899 | 28 | 906 |
| QJR70303.1 | 0.0 | 22 | 899 | 28 | 906 |
| AND19026.1 | 0.0 | 22 | 899 | 28 | 906 |
| AII66187.1 | 0.0 | 22 | 899 | 28 | 906 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3A24_A | 1.28e-199 | 28 | 654 | 6 | 641 | Crystalstructure of BT1871 retaining glycosidase [Bacteroides thetaiotaomicron],3A24_B Crystal structure of BT1871 retaining glycosidase [Bacteroides thetaiotaomicron] |
| 5E1Q_A | 2.30e-198 | 28 | 654 | 20 | 655 | Mutant(D415G) GH97 alpha-galactosidase in complex with Gal-Lac [Bacteroides thetaiotaomicron VPI-5482],5E1Q_B Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac [Bacteroides thetaiotaomicron VPI-5482] |
| 5HQ4_A | 3.74e-56 | 25 | 654 | 2 | 659 | AGlycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8],5HQA_A A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 5HQC_A | 3.74e-56 | 25 | 654 | 2 | 659 | AGlycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 5HQB_A | 9.40e-56 | 25 | 654 | 2 | 659 | AGlycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8A6L0 | 1.10e-203 | 1 | 654 | 1 | 662 | Retaining alpha-galactosidase OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_1871 PE=1 SV=1 |
| G8JZS4 | 1.33e-43 | 2 | 654 | 4 | 724 | Glucan 1,4-alpha-glucosidase SusB OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=susB PE=1 SV=1 |
| D7CFN7 | 1.58e-42 | 24 | 650 | 35 | 617 | Probable retaining alpha-galactosidase OS=Streptomyces bingchenggensis (strain BCW-1) OX=749414 GN=SBI_01652 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
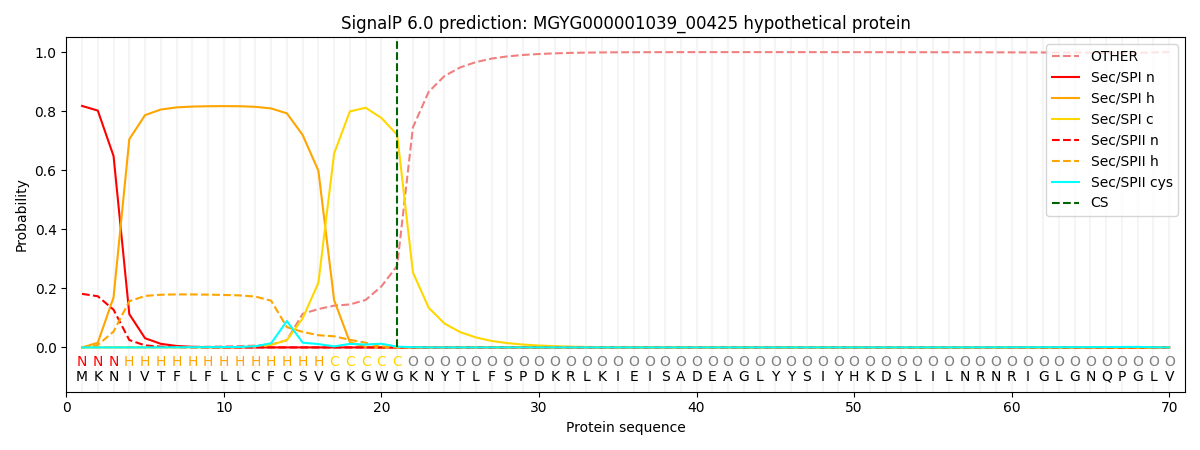
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001983 | 0.805853 | 0.191266 | 0.000320 | 0.000296 | 0.000279 |
