You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001056_00657
You are here: Home > Sequence: MGYG000001056_00657
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
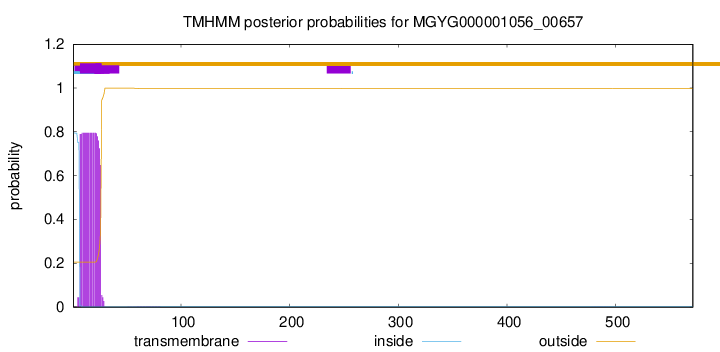
TMHMM annotations
Basic Information help
| Species | Prevotella sp900556395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900556395 | |||||||||||
| CAZyme ID | MGYG000001056_00657 | |||||||||||
| CAZy Family | CE2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24548; End: 26263 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE2 | 148 | 350 | 1.6e-50 | 0.9856459330143541 |
| CE3 | 423 | 564 | 4.9e-16 | 0.6907216494845361 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04501 | SGNH_hydrolase_like_4 | 7.14e-58 | 394 | 567 | 2 | 180 | Members of the SGNH-hydrolase superfamily, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd01831 | Endoglucanase_E_like | 1.15e-33 | 148 | 351 | 2 | 168 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
| cd01828 | sialate_O-acetylesterase_like2 | 3.43e-32 | 394 | 571 | 1 | 169 | sialate_O-acetylesterase_like subfamily of the SGNH-hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 4.09e-29 | 397 | 561 | 1 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd01841 | NnaC_like | 2.11e-21 | 396 | 566 | 4 | 170 | NnaC (CMP-NeuNAc synthetase) _like subfamily of SGNH_hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles two of the three components of typical Ser-His-Asp(Glu) triad from other serine hydrolases. E. coli NnaC appears to be involved in polysaccharide synthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85000.1 | 3.29e-147 | 3 | 359 | 1 | 359 |
| QUB47786.1 | 3.03e-127 | 27 | 360 | 61 | 395 |
| AGY53007.1 | 1.02e-90 | 26 | 357 | 27 | 356 |
| QUT38028.1 | 2.38e-89 | 24 | 357 | 26 | 358 |
| BBL06085.1 | 2.87e-89 | 24 | 359 | 35 | 366 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4IYJ_A | 2.44e-60 | 371 | 566 | 7 | 204 | Crystalstructure of a putative acylhydrolase (BACUNI_03406) from Bacteroides uniformis ATCC 8492 at 1.37 A resolution [Bacteroides uniformis ATCC 8492],4IYJ_B Crystal structure of a putative acylhydrolase (BACUNI_03406) from Bacteroides uniformis ATCC 8492 at 1.37 A resolution [Bacteroides uniformis ATCC 8492] |
| 4PPY_A | 7.05e-58 | 370 | 566 | 3 | 201 | Crystalstructure of a putative acylhydrolase (BF3764) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343],4PPY_B Crystal structure of a putative acylhydrolase (BF3764) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343],4PPY_C Crystal structure of a putative acylhydrolase (BF3764) from Bacteroides fragilis NCTC 9343 at 2.00 A resolution [Bacteroides fragilis NCTC 9343] |
| 4HF7_A | 2.07e-55 | 372 | 566 | 5 | 201 | Crystalstructure of a GDSL-like lipase (BT0569) from Bacteroides thetaiotaomicron VPI-5482 at 1.77 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 3P94_A | 2.14e-50 | 372 | 567 | 5 | 196 | Crystalstructure of a GDSL-like Lipase (BDI_0976) from Parabacteroides distasonis ATCC 8503 at 1.93 A resolution [Parabacteroides distasonis ATCC 8503],3P94_B Crystal structure of a GDSL-like Lipase (BDI_0976) from Parabacteroides distasonis ATCC 8503 at 1.93 A resolution [Parabacteroides distasonis ATCC 8503],3P94_C Crystal structure of a GDSL-like Lipase (BDI_0976) from Parabacteroides distasonis ATCC 8503 at 1.93 A resolution [Parabacteroides distasonis ATCC 8503],3P94_D Crystal structure of a GDSL-like Lipase (BDI_0976) from Parabacteroides distasonis ATCC 8503 at 1.93 A resolution [Parabacteroides distasonis ATCC 8503] |
| 2W9X_A | 2.60e-27 | 37 | 348 | 34 | 350 | Theactive site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus],2W9X_B The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3PDE5 | 1.29e-26 | 37 | 348 | 34 | 350 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ce2C PE=1 SV=1 |
| B3PIB0 | 4.07e-24 | 26 | 357 | 32 | 358 | Acetylxylan esterase / glucomannan deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=axe2C PE=1 SV=1 |
| P10477 | 4.79e-23 | 51 | 357 | 510 | 814 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
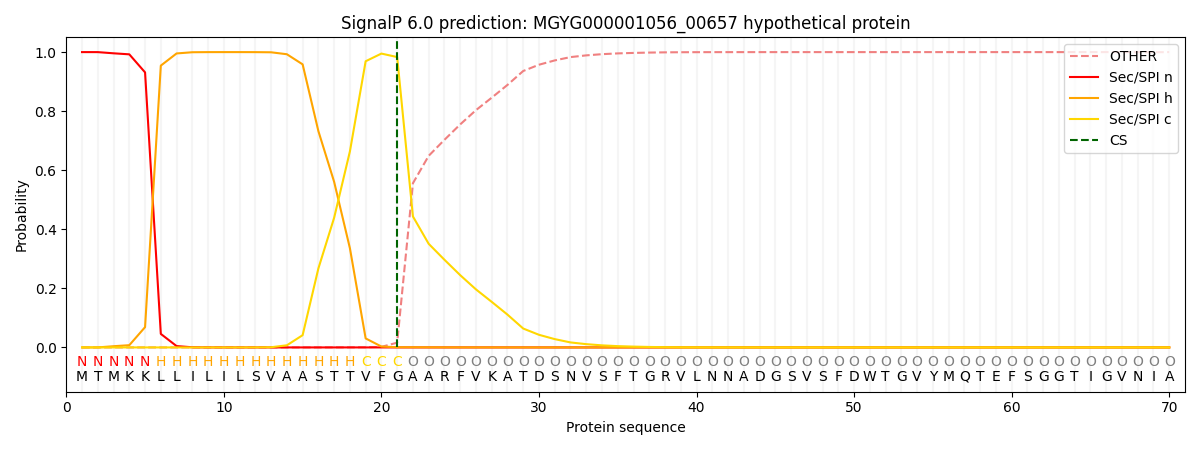
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000494 | 0.998496 | 0.000280 | 0.000252 | 0.000243 | 0.000216 |