You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001063_00067
You are here: Home > Sequence: MGYG000001063_00067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900540475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900540475 | |||||||||||
| CAZyme ID | MGYG000001063_00067 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7232; End: 12604 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 810 | 1122 | 3.3e-63 | 0.9938837920489296 |
| CBM51 | 123 | 268 | 2.4e-34 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08306 | Glyco_hydro_98M | 1.69e-59 | 808 | 1121 | 3 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| pfam08307 | Glyco_hydro_98C | 9.52e-54 | 1124 | 1382 | 1 | 269 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
| pfam08305 | NPCBM | 8.62e-33 | 123 | 268 | 4 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.01e-23 | 123 | 268 | 6 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam07554 | FIVAR | 5.72e-05 | 1533 | 1593 | 1 | 68 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQM60602.1 | 3.99e-134 | 796 | 1380 | 193 | 796 |
| ALG47715.1 | 9.33e-134 | 796 | 1380 | 193 | 792 |
| BAB80035.1 | 5.94e-132 | 796 | 1380 | 193 | 792 |
| AOY52755.1 | 5.94e-132 | 796 | 1380 | 193 | 792 |
| SQG37611.1 | 5.94e-132 | 796 | 1380 | 193 | 792 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WMH_A | 3.14e-116 | 783 | 1380 | 2 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 in complex with the H- disaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMF_A | 3.14e-116 | 783 | 1380 | 2 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
| 2WMI_A | 6.58e-116 | 809 | 1374 | 24 | 598 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_A Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_B Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
| 4D6D_A | 2.57e-115 | 809 | 1374 | 1 | 575 | Crystalstructure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X02 mutant) [Streptococcus pneumoniae SP3-BS71] |
| 2WMI_B | 3.16e-115 | 809 | 1374 | 24 | 598 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6RUF5 | 1.36e-132 | 796 | 1380 | 198 | 797 | Blood-group-substance endo-1,4-beta-galactosidase OS=Clostridium perfringens OX=1502 GN=eabC PE=1 SV=1 |
| P0DTR4 | 1.32e-83 | 477 | 861 | 55 | 457 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| E8MGH9 | 7.21e-13 | 1380 | 1596 | 1656 | 1878 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| Q9L7Q2 | 3.12e-11 | 1390 | 1545 | 416 | 568 | Zinc metalloprotease ZmpB OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=zmpB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
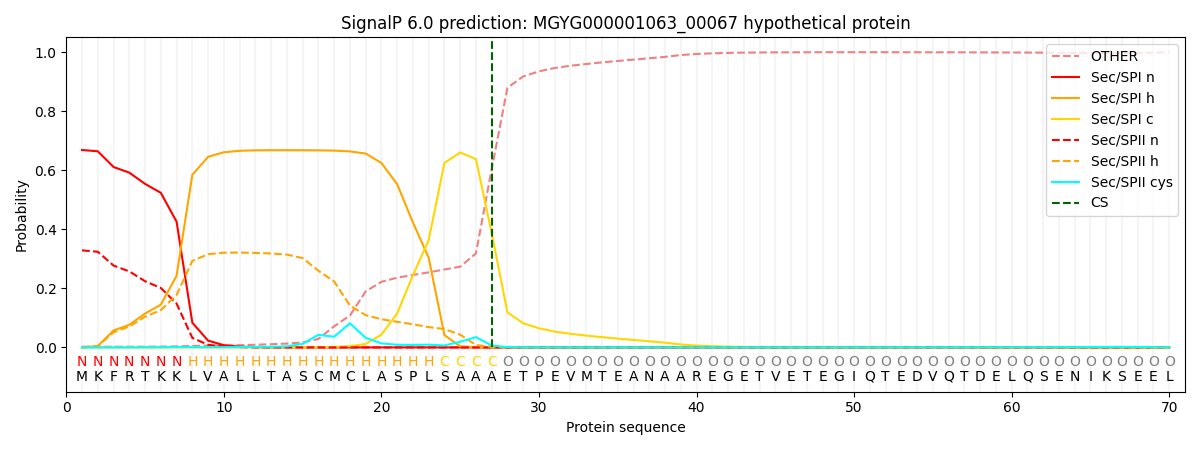
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004077 | 0.655758 | 0.338376 | 0.001033 | 0.000446 | 0.000271 |
