You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001063_02831
You are here: Home > Sequence: MGYG000001063_02831
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900540475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900540475 | |||||||||||
| CAZyme ID | MGYG000001063_02831 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 129422; End: 138721 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 160 | 728 | 3.8e-76 | 0.9905123339658444 |
| GH141 | 1236 | 1721 | 1.2e-53 | 0.9943074003795066 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04084 | CBM6_xylanase-like | 4.65e-22 | 2103 | 2213 | 9 | 118 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| smart00606 | CBD_IV | 2.33e-16 | 2084 | 2215 | 3 | 129 | Cellulose Binding Domain Type IV. |
| pfam03422 | CBM_6 | 1.95e-15 | 2104 | 2213 | 12 | 118 | Carbohydrate binding module (family 6). |
| pfam13229 | Beta_helix | 6.37e-13 | 1509 | 1704 | 2 | 154 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam09479 | Flg_new | 1.72e-12 | 2840 | 2907 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH29314.1 | 7.83e-134 | 155 | 810 | 36 | 683 |
| AUS97976.1 | 3.27e-91 | 1237 | 2107 | 36 | 897 |
| QJD86952.1 | 1.48e-74 | 1237 | 1907 | 7 | 707 |
| AEE97817.1 | 2.11e-73 | 1234 | 2075 | 438 | 1279 |
| QGQ94444.1 | 9.18e-69 | 1231 | 1983 | 45 | 797 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 2.12e-24 | 161 | 467 | 28 | 334 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
| 1UY1_A | 8.96e-10 | 2095 | 2217 | 29 | 143 | Bindingsub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY2_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY3_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY4_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium] |
| 5M77_A | 5.12e-08 | 2099 | 2202 | 391 | 505 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
| 1GMM_A | 9.44e-08 | 2109 | 2210 | 24 | 121 | ChainA, CBM6 [Acetivibrio thermocellus],1UXX_X Chain X, Xylanase U [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33558 | 1.90e-07 | 2095 | 2217 | 258 | 372 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P45796 | 5.09e-07 | 2087 | 2241 | 379 | 540 | Arabinoxylan arabinofuranohydrolase OS=Paenibacillus polymyxa OX=1406 GN=xynD PE=1 SV=1 |
| A0P8X0 | 5.10e-07 | 2916 | 3034 | 815 | 926 | Alpha-amylase OS=Niallia circulans OX=1397 GN=igtZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
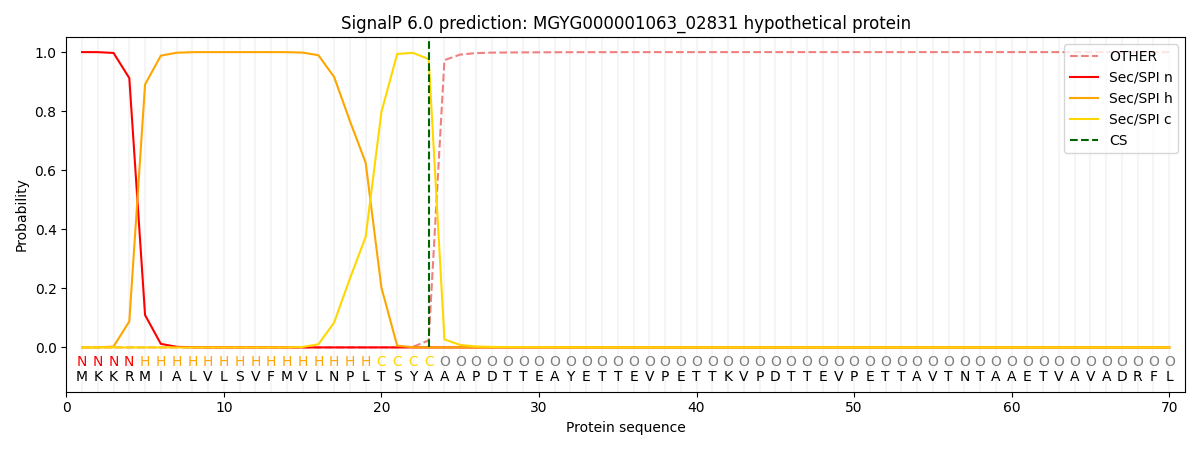
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000696 | 0.998115 | 0.000314 | 0.000312 | 0.000278 | 0.000255 |
