You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001063_04815
You are here: Home > Sequence: MGYG000001063_04815
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900540475 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900540475 | |||||||||||
| CAZyme ID | MGYG000001063_04815 | |||||||||||
| CAZy Family | GH140 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26815; End: 31530 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH140 | 143 | 564 | 8.8e-57 | 0.9927184466019418 |
| CBM32 | 746 | 869 | 1.2e-22 | 0.9435483870967742 |
| CBM32 | 599 | 724 | 3.5e-22 | 0.9354838709677419 |
| CBM32 | 884 | 1007 | 6.8e-20 | 0.9516129032258065 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13204 | DUF4038 | 3.24e-51 | 149 | 468 | 1 | 318 | Protein of unknown function (DUF4038). A family of putative cellulases. |
| pfam16586 | DUF5060 | 1.07e-23 | 44 | 113 | 1 | 70 | Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders. |
| COG5492 | YjdB | 3.71e-18 | 1323 | 1474 | 177 | 321 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam00754 | F5_F8_type_C | 1.08e-16 | 887 | 1010 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.18e-15 | 597 | 724 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK56434.1 | 7.67e-139 | 17 | 1011 | 8 | 962 |
| QTH41603.1 | 2.30e-106 | 5 | 1011 | 3 | 1164 |
| BBI36498.1 | 6.94e-95 | 48 | 1012 | 41 | 1051 |
| QTH45264.1 | 1.62e-90 | 44 | 573 | 39 | 573 |
| AZM49602.1 | 1.71e-83 | 1 | 568 | 1 | 569 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MSY_A | 4.71e-21 | 141 | 537 | 15 | 414 | Glycosidehydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482],5MSY_B Glycoside hydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482],5MSY_C Glycoside hydrolase BT_1012 [Bacteroides thetaiotaomicron VPI-5482] |
| 3KZS_A | 6.19e-19 | 141 | 477 | 15 | 356 | Crystalstructure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_B Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_C Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482],3KZS_D Crystal structure of glycosyl hydrolase family 5 (NP_809925.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 2.10 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4QFU_A | 2.24e-18 | 140 | 537 | 27 | 430 | ChainA, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_B Chain B, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_C Chain C, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_D Chain D, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_E Chain E, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_F Chain F, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_G Chain G, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_H Chain H, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_I Chain I, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_J Chain J, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_K Chain K, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482],4QFU_L Chain L, glycoside hydrolase family 5 [Phocaeicola vulgatus ATCC 8482] |
| 4N0R_A | 5.41e-09 | 44 | 327 | 4 | 282 | ChainA, putative glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],4N0R_B Chain B, putative glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33747 | 3.74e-11 | 1323 | 1486 | 33 | 199 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P35838 | 2.84e-08 | 1332 | 1412 | 241 | 321 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
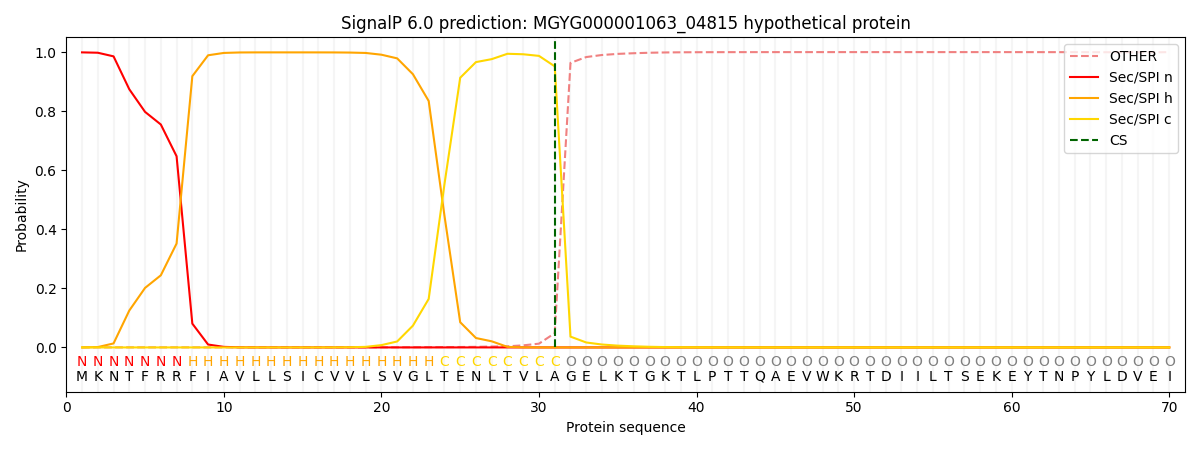
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001931 | 0.995729 | 0.001319 | 0.000430 | 0.000319 | 0.000261 |

