You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001067_01837
You are here: Home > Sequence: MGYG000001067_01837
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1590 sp900553245 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_B; Peptococcia; Peptococcales; Peptococcaceae; UMGS1590; UMGS1590 sp900553245 | |||||||||||
| CAZyme ID | MGYG000001067_01837 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2209; End: 3837 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 332 | 452 | 3.4e-27 | 0.36303630363036304 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 5.47e-14 | 290 | 506 | 113 | 328 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 2.22e-09 | 326 | 442 | 129 | 243 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB68979.1 | 3.41e-121 | 63 | 536 | 36 | 505 |
| AFS79156.1 | 1.35e-120 | 61 | 538 | 52 | 526 |
| QUH19438.1 | 3.79e-120 | 71 | 536 | 59 | 523 |
| QHI71412.1 | 7.63e-120 | 44 | 536 | 18 | 507 |
| QOX62356.1 | 4.85e-119 | 70 | 536 | 47 | 510 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVD_A | 2.77e-33 | 298 | 536 | 56 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 2.77e-33 | 298 | 536 | 56 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 3.25e-33 | 298 | 536 | 56 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 7.05e-33 | 298 | 536 | 56 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 6ZPL_C | 1.49e-32 | 298 | 536 | 48 | 272 | ChainC, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 4.09e-31 | 298 | 542 | 78 | 308 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| P16699 | 1.80e-06 | 348 | 436 | 185 | 271 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
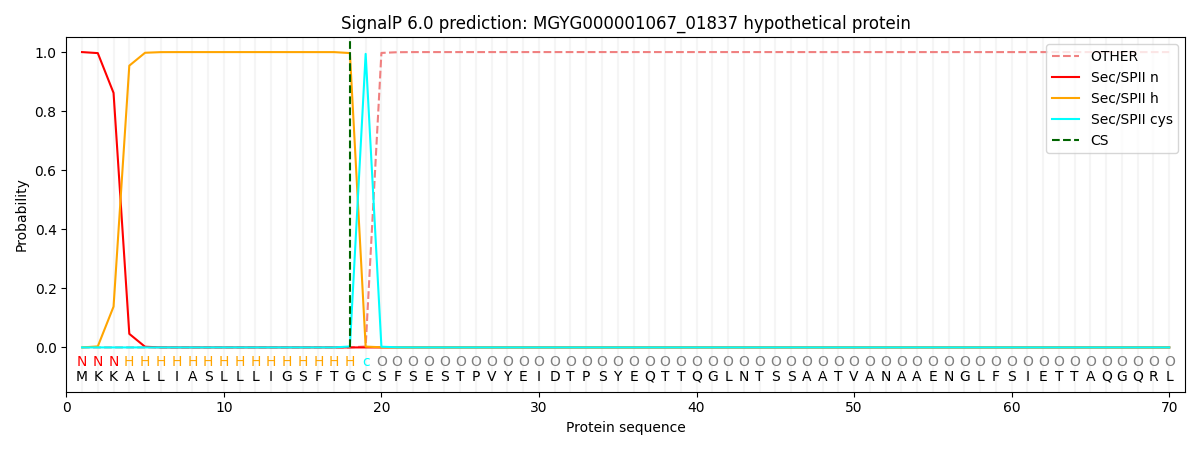
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000046 | 0.000000 | 0.000000 | 0.000000 |
