You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001073_00886
You are here: Home > Sequence: MGYG000001073_00886
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusobacterium_B sp900554885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium_B; Fusobacterium_B sp900554885 | |||||||||||
| CAZyme ID | MGYG000001073_00886 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 316; End: 2424 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 32 | 371 | 1e-51 | 0.9649122807017544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 4.57e-45 | 41 | 373 | 14 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13859 | BNR_3 | 2.60e-10 | 52 | 279 | 9 | 212 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13088 | BNR_2 | 2.29e-08 | 75 | 282 | 20 | 202 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| smart00869 | Autotransporter | 1.74e-06 | 429 | 693 | 4 | 266 | Autotransporter beta-domain. Secretion of protein products occurs by a number of different pathways in bacteria. One of these pathways known as the type IV pathway was first described for the IgA1 protease. The protein component that mediates secretion through the outer membrane is contained within the secreted protein itself, hence the proteins secreted in this way are called autotransporters. This family corresponds to the presumed integral membrane beta-barrel domain that transports the protein. This domain is found at the C-terminus of the proteins it occurs in. The N-terminus contains the variable passenger domain that is translocated across the membrane. Once the passenger domain is exported it is cleaved auto-catalytically in some proteins, in others a different peptidase is used and in some cases no cleavage occurs. |
| pfam03797 | Autotransporter | 1.41e-05 | 470 | 685 | 44 | 254 | Autotransporter beta-domain. Secretion of protein products occurs by a number of different pathways in bacteria. One of these pathways known as the type V pathway was first described for the IgA1 protease. The protein component that mediates secretion through the outer membrane is contained within the secreted protein itself, hence the proteins secreted in this way are called autotransporters. This family corresponds to the presumed integral membrane beta-barrel domain that transports the protein. This domain is found at the C-terminus of the proteins it occurs in. The N-terminus contains the variable passenger domain that is translocated across the membrane. Once the passenger domain is exported it is cleaved auto-catalytically in some proteins, in others a different protease is used and in some cases no cleavage occurs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM15078.1 | 8.32e-235 | 1 | 702 | 1 | 715 |
| AMZ14174.1 | 5.90e-180 | 21 | 700 | 8 | 677 |
| AVQ18134.1 | 3.73e-168 | 231 | 702 | 8 | 464 |
| QPR26713.1 | 1.85e-161 | 21 | 700 | 32 | 701 |
| ASG76455.1 | 7.22e-161 | 97 | 700 | 37 | 623 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MYV_A | 1.96e-18 | 37 | 363 | 177 | 508 | Sialidase26co-crystallized with DANA-Gc [bacterium],6MYV_B Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_C Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_D Sialidase26 co-crystallized with DANA-Gc [bacterium] |
| 6MRV_A | 2.17e-18 | 37 | 363 | 198 | 529 | Sialidase26co-crystallized with DANA [bacterium],6MRV_B Sialidase26 co-crystallized with DANA [bacterium],6MRX_A Sialidase26 apo [bacterium],6MRX_B Sialidase26 apo [bacterium],6MRX_C Sialidase26 apo [bacterium],6MRX_D Sialidase26 apo [bacterium] |
| 4FJ6_A | 2.57e-18 | 37 | 363 | 177 | 508 | Crystalstructure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_B Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_C Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_D Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503] |
| 4Q6K_A | 1.87e-17 | 37 | 363 | 180 | 511 | Crystalstructure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_B Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_C Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_D Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185] |
| 1EUR_A | 2.88e-15 | 33 | 284 | 17 | 260 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31206 | 1.25e-19 | 41 | 363 | 203 | 530 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
| P15698 | 1.61e-15 | 29 | 279 | 41 | 283 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
| Q02834 | 4.87e-14 | 33 | 284 | 59 | 302 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P10481 | 1.83e-12 | 41 | 282 | 36 | 268 | Sialidase OS=Clostridium perfringens OX=1502 GN=nanH PE=1 SV=1 |
| P29768 | 1.01e-10 | 28 | 379 | 22 | 381 | Sialidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=nanH PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
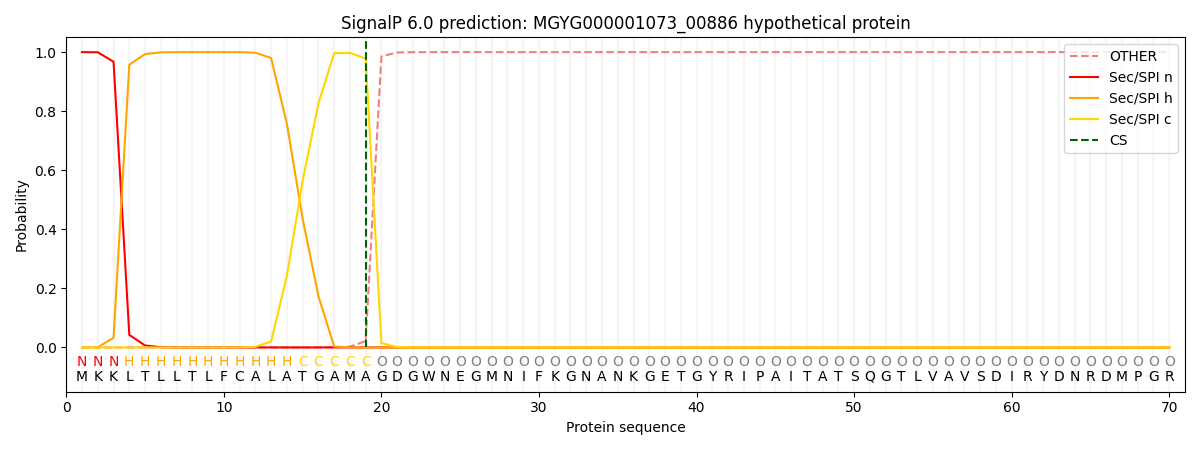
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000299 | 0.998806 | 0.000274 | 0.000199 | 0.000192 | 0.000187 |
