You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001081_01378
You are here: Home > Sequence: MGYG000001081_01378
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
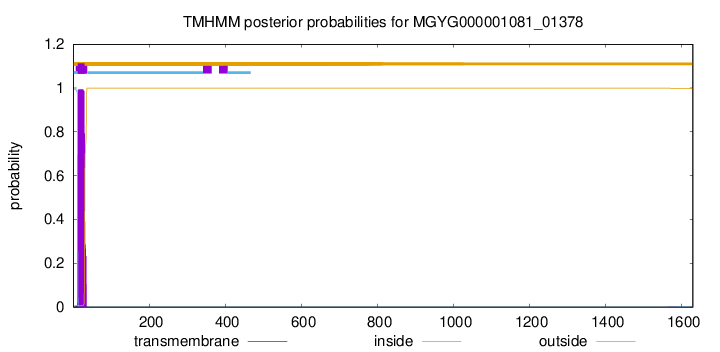
TMHMM annotations
Basic Information help
| Species | Acutalibacter sp900755895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Acutalibacter; Acutalibacter sp900755895 | |||||||||||
| CAZyme ID | MGYG000001081_01378 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66651; End: 71543 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 196 | 313 | 1.6e-23 | 0.9736842105263158 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00395 | SLH | 4.05e-09 | 150 | 191 | 1 | 41 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 3.05e-07 | 31 | 200 | 583 | 752 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 4.33e-07 | 89 | 130 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 1.96e-05 | 89 | 198 | 582 | 689 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 0.002 | 33 | 71 | 4 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQR07714.1 | 0.0 | 10 | 1593 | 10 | 1581 |
| QIA32369.1 | 0.0 | 10 | 1593 | 10 | 1581 |
| BCK84769.1 | 0.0 | 10 | 1528 | 17 | 1540 |
| QQR30851.1 | 1.32e-237 | 1 | 1427 | 1 | 1588 |
| ASB41592.1 | 1.32e-237 | 1 | 1427 | 1 | 1588 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 5.50e-08 | 33 | 198 | 29 | 194 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 5.58e-08 | 33 | 198 | 8 | 173 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 5GZT_A | 1.11e-06 | 217 | 267 | 58 | 108 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 5.93e-20 | 37 | 203 | 1688 | 1861 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38537 | 2.83e-18 | 5 | 232 | 3 | 238 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P38535 | 4.08e-17 | 37 | 196 | 914 | 1077 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q06852 | 1.72e-13 | 52 | 200 | 2104 | 2264 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
| P19424 | 2.56e-12 | 88 | 206 | 40 | 154 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
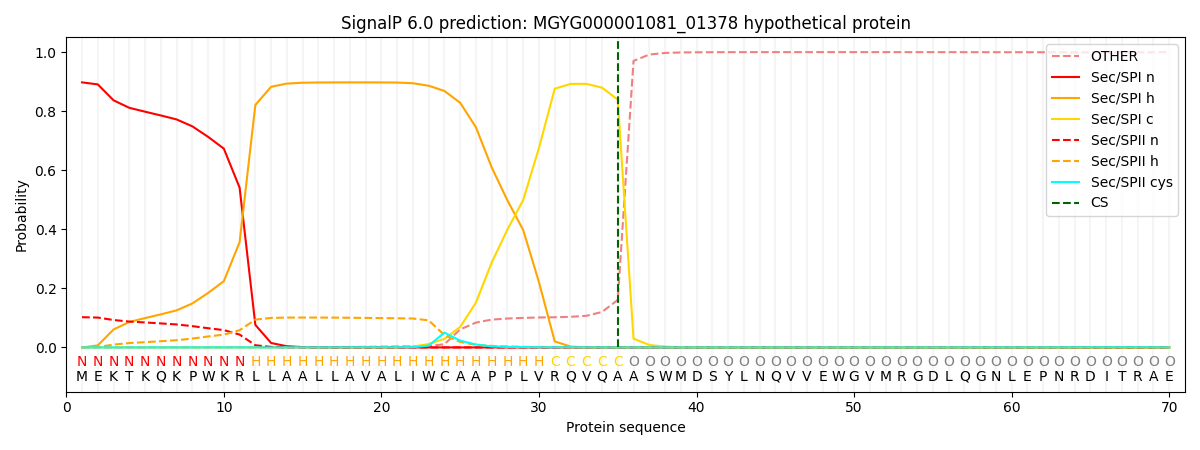
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000912 | 0.890608 | 0.107579 | 0.000308 | 0.000293 | 0.000262 |