You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001082_02162
You are here: Home > Sequence: MGYG000001082_02162
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
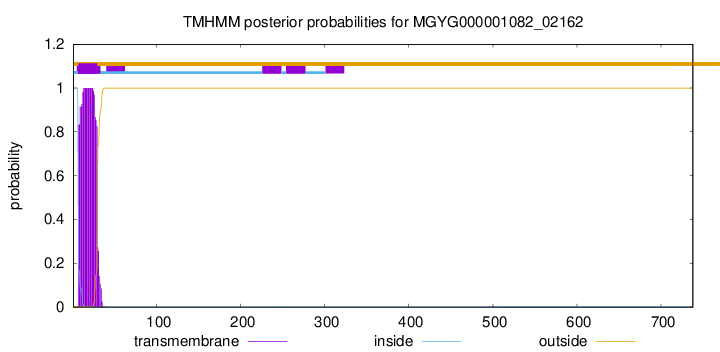
TMHMM annotations
Basic Information help
| Species | Ruminococcus_E sp900755995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Ruminococcus_E; Ruminococcus_E sp900755995 | |||||||||||
| CAZyme ID | MGYG000001082_02162 | |||||||||||
| CAZy Family | CBM74 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6072; End: 8288 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM74 | 243 | 569 | 9.9e-130 | 0.9967948717948718 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16738 | CBM26 | 3.76e-11 | 39 | 109 | 1 | 68 | Starch-binding module 26. CBM26 is a carbohydrate-binding module that binds starch. |
| cd14256 | Dockerin_I | 2.50e-08 | 676 | 738 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| PRK14081 | PRK14081 | 2.40e-05 | 587 | 674 | 293 | 385 | triple tyrosine motif-containing protein; Provisional |
| pfam00404 | Dockerin_1 | 4.72e-04 | 677 | 737 | 1 | 55 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam07495 | Y_Y_Y | 0.001 | 609 | 666 | 2 | 64 | Y_Y_Y domain. This domain is mostly found at the end of the beta propellers (pfam07494) in a family of two component regulators. However they are also found tandemly repeated in CTC_02402 without other signal conduction domains being present. It's named after the conserved tyrosines found in the alignment. The exact function is not known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT06902.1 | 0.0 | 1 | 697 | 1 | 693 |
| CDM67953.1 | 1.48e-107 | 48 | 657 | 583 | 1172 |
| AWX97480.1 | 9.12e-86 | 36 | 705 | 551 | 1191 |
| AWX95531.1 | 9.12e-86 | 36 | 705 | 551 | 1191 |
| QCT07491.1 | 1.36e-72 | 1 | 573 | 1 | 528 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.049103 | 0.936059 | 0.013168 | 0.000821 | 0.000422 | 0.000397 |