You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001085_00286
You are here: Home > Sequence: MGYG000001085_00286
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000434935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000434935 | |||||||||||
| CAZyme ID | MGYG000001085_00286 | |||||||||||
| CAZy Family | GH158 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43235; End: 44566 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH158 | 53 | 185 | 4e-45 | 0.9848484848484849 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02836 | Glyco_hydro_2_C | 3.49e-17 | 74 | 326 | 46 | 299 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 1.14e-12 | 74 | 240 | 323 | 510 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 3.26e-05 | 41 | 217 | 286 | 457 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 6.44e-04 | 121 | 179 | 416 | 477 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 0.002 | 128 | 202 | 436 | 503 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU95349.1 | 8.68e-162 | 4 | 443 | 6 | 451 |
| QNL37133.1 | 8.68e-162 | 19 | 443 | 19 | 451 |
| QDH55227.1 | 5.71e-160 | 19 | 443 | 19 | 451 |
| QEH43942.1 | 2.46e-154 | 12 | 443 | 15 | 453 |
| QNL47842.1 | 1.30e-134 | 6 | 443 | 4 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5C70_A | 4.80e-08 | 77 | 240 | 332 | 515 | Thestructure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae],5C70_B The structure of Aspergillus oryzae beta-glucuronidase [Aspergillus oryzae] |
| 1BHG_A | 6.36e-08 | 73 | 215 | 353 | 485 | HumanBeta-Glucuronidase At 2.6 A Resolution [Homo sapiens],1BHG_B Human Beta-Glucuronidase At 2.6 A Resolution [Homo sapiens],3HN3_A Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_B Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_D Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_E Human beta-glucuronidase at 1.7 A resolution [Homo sapiens] |
| 5C71_A | 2.62e-07 | 77 | 240 | 357 | 540 | Thestructure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_B The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_C The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae],5C71_D The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide [Aspergillus oryzae] |
| 6LEM_B | 7.77e-07 | 77 | 215 | 320 | 467 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
| 6LEJ_B | 7.78e-07 | 77 | 215 | 322 | 469 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O97524 | 2.05e-07 | 73 | 215 | 372 | 504 | Beta-glucuronidase OS=Felis catus OX=9685 GN=GUSB PE=1 SV=1 |
| Q5R5N6 | 2.05e-07 | 73 | 215 | 373 | 505 | Beta-glucuronidase OS=Pongo abelii OX=9601 GN=GUSB PE=2 SV=2 |
| P08236 | 3.58e-07 | 73 | 215 | 373 | 505 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| O77695 | 4.73e-07 | 73 | 215 | 370 | 502 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
| O18835 | 4.74e-07 | 73 | 215 | 372 | 504 | Beta-glucuronidase OS=Canis lupus familiaris OX=9615 GN=GUSB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
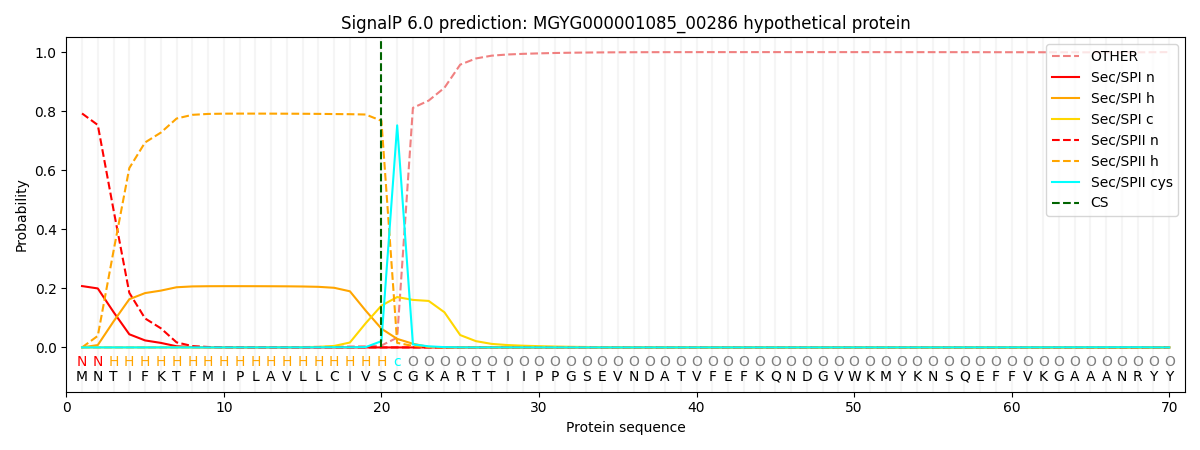
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000210 | 0.203958 | 0.795559 | 0.000092 | 0.000100 | 0.000087 |
