You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001091_01559
You are here: Home > Sequence: MGYG000001091_01559
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Citrobacter_A sp009363175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Citrobacter_A; Citrobacter_A sp009363175 | |||||||||||
| CAZyme ID | MGYG000001091_01559 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | Putative acyl-CoA thioester hydrolase YbhC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35; End: 1270 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10531 | PRK10531 | 0.0 | 2 | 404 | 18 | 422 | putative acyl-CoA thioester hydrolase. |
| COG4677 | PemB | 0.0 | 1 | 405 | 17 | 405 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02708 | PLN02708 | 1.76e-09 | 53 | 290 | 216 | 413 | Probable pectinesterase/pectinesterase inhibitor |
| pfam01095 | Pectinesterase | 3.35e-08 | 65 | 403 | 1 | 266 | Pectinesterase. |
| PLN02301 | PLN02301 | 7.89e-08 | 40 | 299 | 205 | 416 | pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFS72645.1 | 8.90e-298 | 1 | 411 | 17 | 427 |
| AMG93924.1 | 9.95e-295 | 1 | 411 | 17 | 427 |
| AUO65165.1 | 1.41e-294 | 1 | 411 | 17 | 427 |
| QZE46353.1 | 2.01e-294 | 1 | 411 | 17 | 427 |
| AKE58269.1 | 2.85e-294 | 1 | 411 | 17 | 427 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GRH_A | 6.21e-250 | 16 | 411 | 27 | 422 | Crystalstructure of escherichia coli ybhc [Escherichia coli K-12] |
| 4PMH_A | 4.68e-65 | 17 | 403 | 1 | 366 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P46130 | 2.01e-256 | 1 | 411 | 17 | 427 | Putative acyl-CoA thioester hydrolase YbhC OS=Escherichia coli (strain K12) OX=83333 GN=ybhC PE=1 SV=2 |
| Q47474 | 3.44e-113 | 3 | 397 | 19 | 426 | Pectinesterase B OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemB PE=1 SV=2 |
| P55743 | 1.77e-110 | 19 | 398 | 5 | 396 | Pectinesterase B OS=Pectobacterium parmentieri OX=1905730 GN=pemB PE=3 SV=2 |
| E7CIP7 | 2.58e-65 | 15 | 403 | 15 | 382 | Pectinesterase OS=Sitophilus oryzae OX=7048 GN=CE8-1 PE=1 SV=1 |
| P58601 | 5.08e-29 | 23 | 403 | 30 | 394 | Pectinesterase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=pme PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
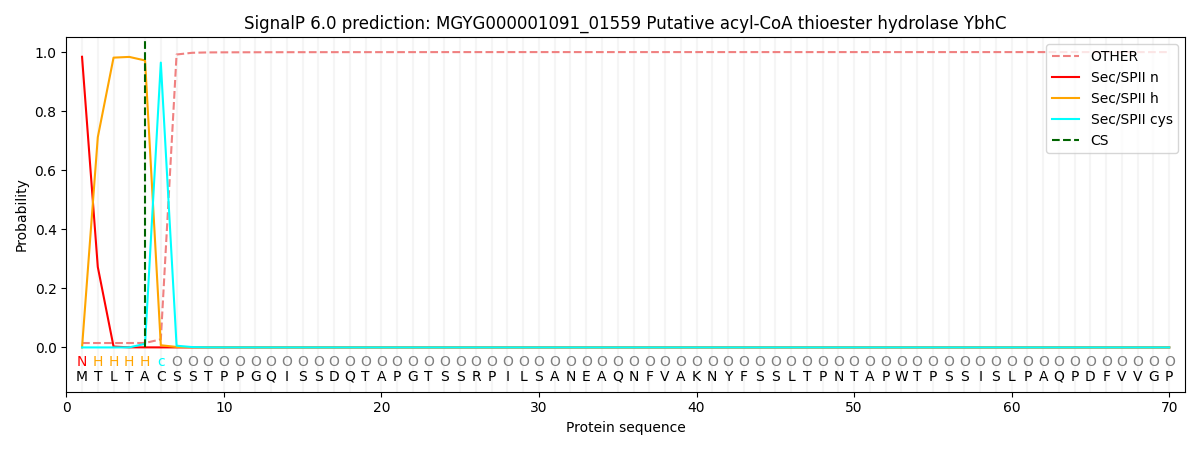
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014914 | 0.000900 | 0.984212 | 0.000001 | 0.000002 | 0.000001 |
