You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001091_03638
You are here: Home > Sequence: MGYG000001091_03638
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
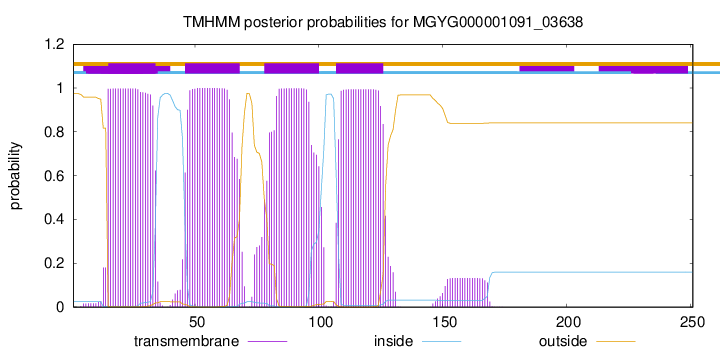
TMHMM annotations
Basic Information help
| Species | Citrobacter_A sp009363175 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Citrobacter_A; Citrobacter_A sp009363175 | |||||||||||
| CAZyme ID | MGYG000001091_03638 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3118; End: 3873 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 1 | 222 | 6.1e-47 | 0.42777777777777776 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13279 | arnT | 5.40e-118 | 1 | 250 | 302 | 552 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1807 | ArnT | 3.70e-28 | 1 | 224 | 305 | 535 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFS71066.1 | 2.74e-176 | 1 | 251 | 301 | 551 |
| QZE47906.1 | 5.43e-174 | 1 | 251 | 313 | 563 |
| AKE59276.1 | 2.09e-173 | 1 | 251 | 301 | 551 |
| QZA35078.1 | 1.13e-170 | 1 | 251 | 301 | 551 |
| SAZ37024.1 | 1.13e-170 | 1 | 251 | 301 | 551 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1LLL1 | 2.01e-134 | 1 | 251 | 300 | 550 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia coli (strain SMS-3-5 / SECEC) OX=439855 GN=arnT PE=3 SV=1 |
| B5YXQ0 | 2.01e-134 | 1 | 251 | 300 | 550 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=arnT PE=3 SV=1 |
| B7LAS2 | 2.01e-134 | 1 | 251 | 300 | 550 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia coli (strain 55989 / EAEC) OX=585055 GN=arnT PE=3 SV=1 |
| Q8XDZ1 | 2.01e-134 | 1 | 251 | 300 | 550 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia coli O157:H7 OX=83334 GN=arnT PE=3 SV=1 |
| Q0TFI5 | 4.03e-134 | 1 | 251 | 300 | 550 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Escherichia coli O6:K15:H31 (strain 536 / UPEC) OX=362663 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
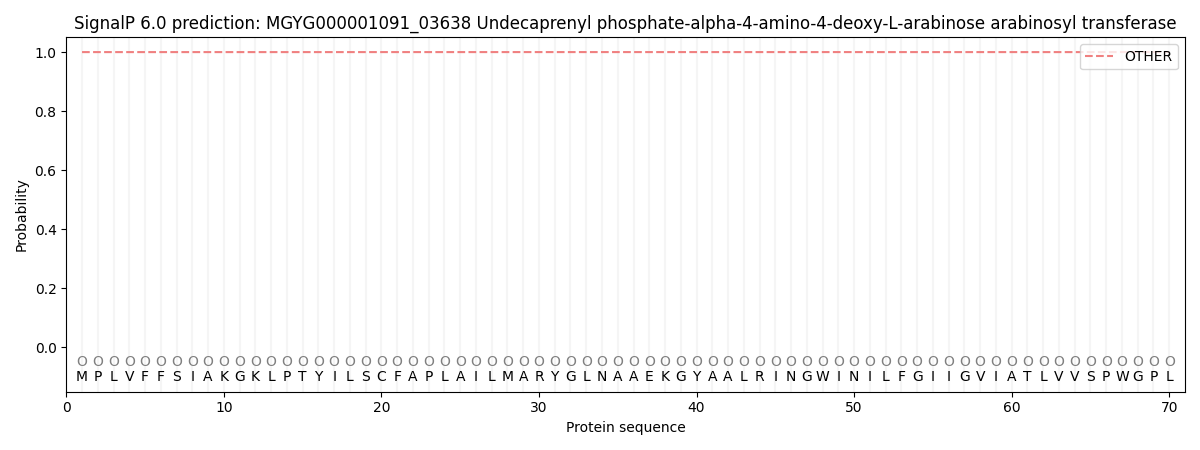
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000021 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |