You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001094_01055
You are here: Home > Sequence: MGYG000001094_01055
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Azonexus sp900549295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Rhodocyclaceae; Azonexus; Azonexus sp900549295 | |||||||||||
| CAZyme ID | MGYG000001094_01055 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12759; End: 15560 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 47 | 267 | 2.9e-79 | 0.9955357142857143 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10922 | CE4_PelA_like_C | 2.64e-133 | 498 | 764 | 1 | 266 | C-terminal Putative NodB-like catalytic domain of PelA-like uncharacterized hypothetical proteins found in bacteria. This family is represented by a protein PelA of unknown function that is encoded by a gene in the pelA-G gene cluster for pellicle production and biofilm formation in Pseudomonas aeruginosa. PelA and most of the family members contain a domain of unknown function, DUF297, in the N-terminus and a C-terminal domain that shows high sequence similarity to the catalytic domain of the six-stranded barrel rhizobial NodB-like proteins, which remove N-linked or O-linked acetyl groups from cell wall polysaccharides and belong to the larger carbohydrate esterase 4 (CE4) superfamily. |
| COG3868 | COG3868 | 3.34e-59 | 11 | 335 | 5 | 305 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
| pfam03537 | Glyco_hydro_114 | 5.80e-19 | 29 | 267 | 3 | 219 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG2342 | COG2342 | 9.91e-11 | 46 | 268 | 44 | 285 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| cd10585 | CE4_SF | 3.24e-08 | 502 | 647 | 1 | 128 | Catalytic NodB homology domain of the carbohydrate esterase 4 superfamily. The carbohydrate esterase 4 (CE4) superfamily mainly includes chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan, respectively. Members in this superfamily contain a NodB homology domain that adopts a deformed (beta/alpha)8 barrel fold, which encompasses a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad, closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. The NodB homology domain of CE4 superfamily is remotely related to the 7-stranded beta/alpha barrel catalytic domain of the superfamily consisting of family 38 glycoside hydrolases (GH38), family 57 heat stable retaining glycoside hydrolases (GH57), lactam utilization protein LamB/YcsF family proteins, and YdjC-family proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRM20898.1 | 0.0 | 21 | 932 | 16 | 926 |
| ABM31640.1 | 0.0 | 14 | 931 | 5 | 938 |
| ATG95274.1 | 0.0 | 14 | 931 | 5 | 938 |
| QCX11560.1 | 0.0 | 14 | 931 | 5 | 938 |
| AVT12040.1 | 0.0 | 14 | 931 | 5 | 947 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5TCB_A | 1.99e-46 | 28 | 279 | 25 | 278 | Structureof the glycoside hydrolase domain of PelA from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5TSY_A | 1.28e-45 | 28 | 279 | 25 | 278 | Structureof the glycoside hydrolase domain of PelA variant E218A from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
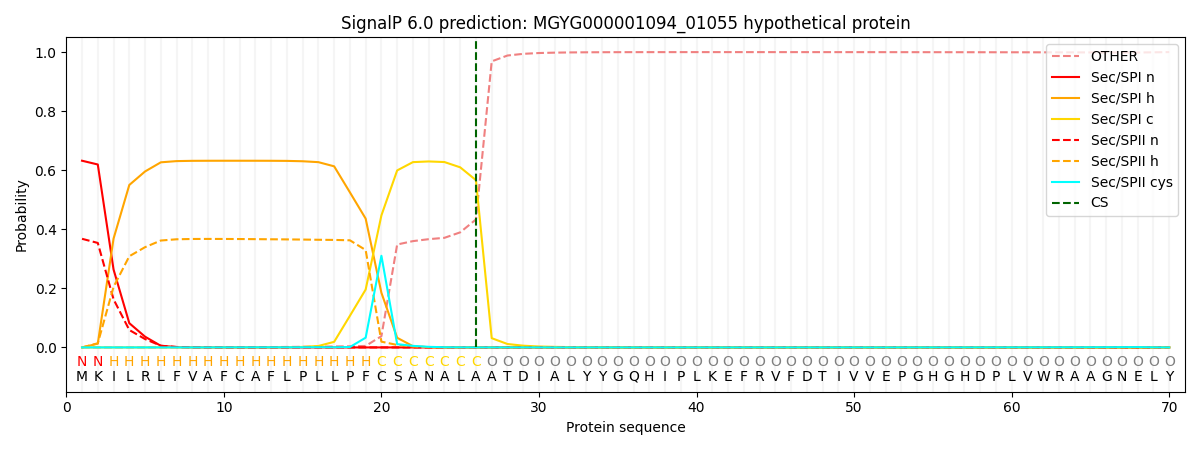
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000578 | 0.624637 | 0.374078 | 0.000283 | 0.000221 | 0.000191 |
