You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001095_00580
You are here: Home > Sequence: MGYG000001095_00580
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Chryseobacterium gambrini | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Flavobacteriales; Weeksellaceae; Chryseobacterium; Chryseobacterium gambrini | |||||||||||
| CAZyme ID | MGYG000001095_00580 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2772; End: 4043 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 62 | 373 | 1.4e-28 | 0.9421221864951769 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5309 | Scw11 | 1.67e-18 | 29 | 366 | 35 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| pfam00332 | Glyco_hydro_17 | 1.24e-10 | 74 | 373 | 17 | 307 | Glycosyl hydrolases family 17. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VFB04068.1 | 6.81e-311 | 1 | 423 | 1 | 423 |
| QQV02671.1 | 6.81e-311 | 1 | 423 | 1 | 423 |
| QZO83107.1 | 5.55e-216 | 1 | 411 | 1 | 413 |
| QDE20671.1 | 1.06e-213 | 1 | 411 | 1 | 413 |
| QWU97811.1 | 1.06e-213 | 1 | 411 | 1 | 413 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FCG_A | 3.00e-192 | 7 | 411 | 9 | 421 | Crystalstructure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_B Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_C Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_D Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_E Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131],6FCG_F Crystal structure of an endo-laminarinase from Formosa Hel1_33_131 [Formosa sp. Hel1_33_131] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QN74 | 7.27e-11 | 56 | 373 | 400 | 673 | Putative glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=btgC PE=3 SV=2 |
| Q0CI96 | 1.58e-09 | 52 | 370 | 367 | 641 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgC PE=3 SV=2 |
| D4B2W4 | 5.02e-09 | 291 | 371 | 216 | 297 | Glucan 1,3-beta-glucosidase ARB_02797 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02797 PE=1 SV=1 |
| B8NTP7 | 3.56e-08 | 17 | 348 | 352 | 647 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgC PE=3 SV=2 |
| Q2U492 | 3.56e-08 | 17 | 348 | 352 | 647 | Probable glucan endo-1,3-beta-glucosidase btgC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
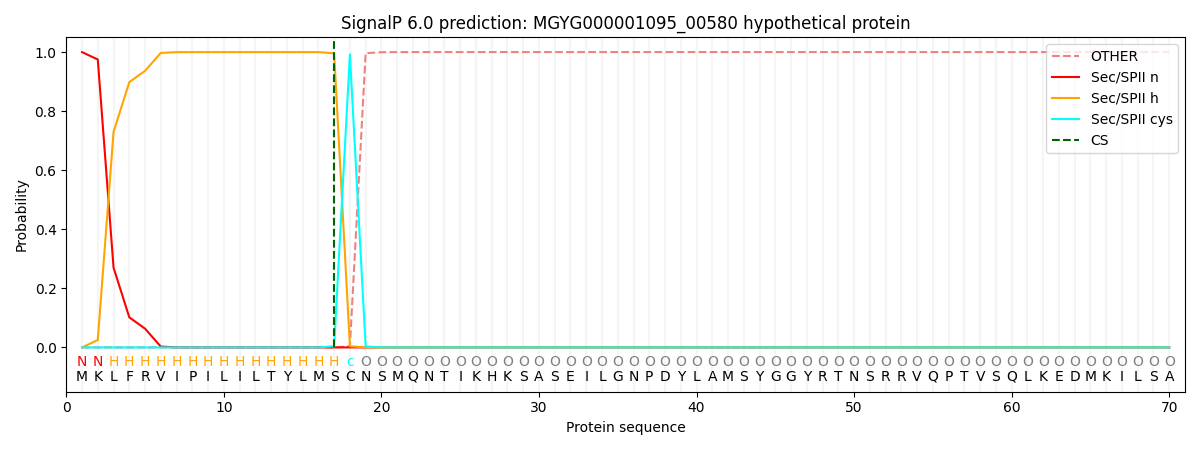
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000053 | 0.000000 | 0.000000 | 0.000000 |
