You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001095_01100
You are here: Home > Sequence: MGYG000001095_01100
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
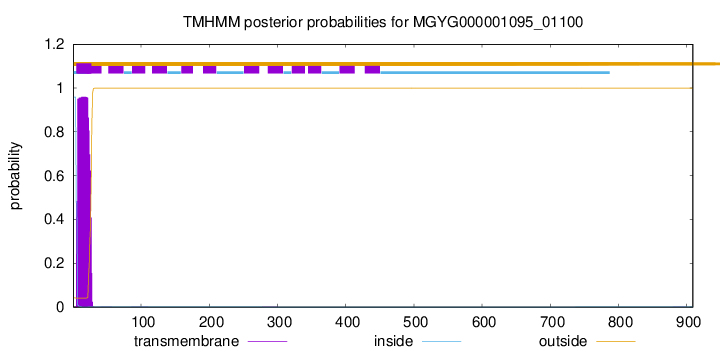
TMHMM annotations
Basic Information help
| Species | Chryseobacterium gambrini | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Flavobacteriales; Weeksellaceae; Chryseobacterium; Chryseobacterium gambrini | |||||||||||
| CAZyme ID | MGYG000001095_01100 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5404; End: 8133 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 27 | 383 | 3.3e-115 | 0.9824561403508771 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 1.04e-54 | 1 | 429 | 5 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam18962 | Por_Secre_tail | 2.14e-11 | 837 | 907 | 1 | 71 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
| TIGR04183 | Por_Secre_tail | 1.76e-09 | 837 | 909 | 1 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam02055 | Glyco_hydro_30 | 2.77e-06 | 73 | 296 | 65 | 299 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam02057 | Glyco_hydro_59 | 5.88e-05 | 135 | 290 | 101 | 241 | Glycosyl hydrolase family 59. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWA38801.1 | 0.0 | 1 | 909 | 1 | 909 |
| AZA86004.1 | 0.0 | 1 | 908 | 1 | 906 |
| AZA94412.1 | 0.0 | 1 | 908 | 1 | 906 |
| AZA57764.1 | 0.0 | 1 | 908 | 1 | 906 |
| QWT87024.1 | 0.0 | 1 | 909 | 1 | 912 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1NOF_A | 2.68e-55 | 27 | 427 | 2 | 381 | ChainA, xylanase [Dickeya chrysanthemi],2Y24_A Chain A, XYLANASE [Dickeya chrysanthemi] |
| 4CKQ_A | 4.50e-51 | 23 | 427 | 19 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 2.91e-50 | 23 | 427 | 19 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
| 4UQA_A | 2.91e-50 | 23 | 427 | 19 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 3GTN_A | 1.39e-46 | 28 | 430 | 5 | 391 | CrystalStructure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3GTN_B Crystal Structure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3KL0_A Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_B Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_C Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_D Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL3_A Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_B Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_C Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_D Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL5_A Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_B Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_C Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_D Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45070 | 1.22e-45 | 28 | 430 | 36 | 422 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 1.42e-42 | 29 | 418 | 38 | 400 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| G2Q1N4 | 2.04e-06 | 26 | 374 | 27 | 414 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
| Q8J0I9 | 2.75e-06 | 31 | 405 | 69 | 486 | Endo-1,6-beta-D-glucanase BGN16.3 OS=Trichoderma harzianum OX=5544 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
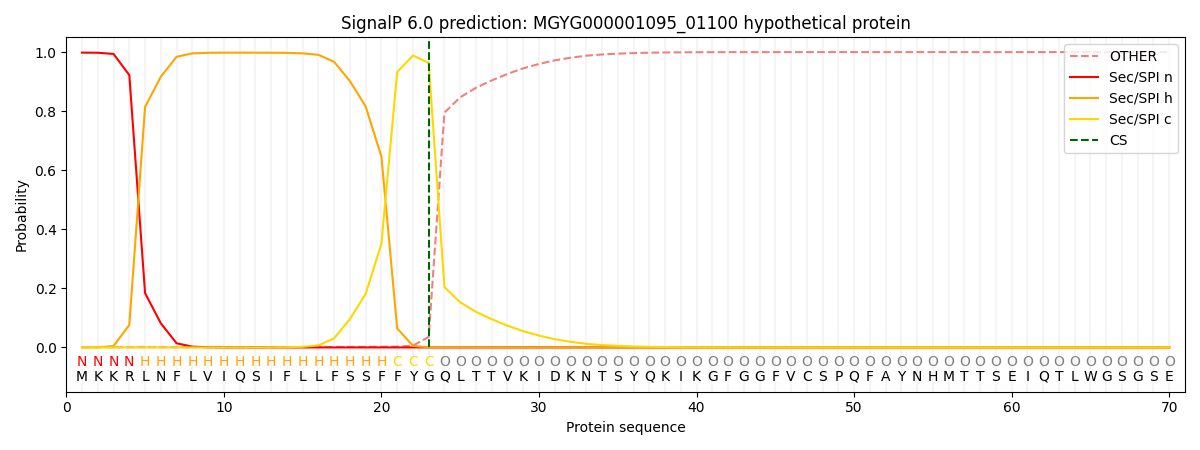
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002276 | 0.995727 | 0.001330 | 0.000233 | 0.000191 | 0.000189 |