You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001124_01201
You are here: Home > Sequence: MGYG000001124_01201
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
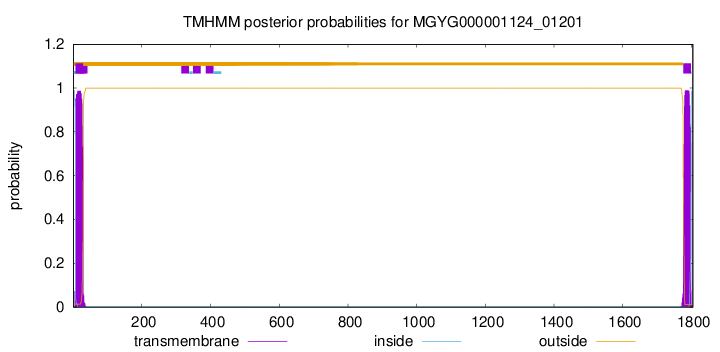
TMHMM annotations
Basic Information help
| Species | Collinsella sp900757285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900757285 | |||||||||||
| CAZyme ID | MGYG000001124_01201 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 567; End: 5981 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 115 | 244 | 8.8e-19 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09479 | Flg_new | 1.39e-14 | 1664 | 1726 | 2 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| pfam00754 | F5_F8_type_C | 1.21e-13 | 110 | 244 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13385 | Laminin_G_3 | 2.30e-08 | 1220 | 1337 | 17 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| TIGR02543 | List_Bact_rpt | 2.62e-06 | 1691 | 1729 | 1 | 41 | Listeria/Bacterioides repeat. This model describes a conserved core region, about 43 residues in length, of at least two families of tandem repeats. These include 78-residue repeats from 2 to 15 in number, in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few bacteria. [Unknown function, General] |
| cd14791 | GH36 | 1.12e-05 | 627 | 812 | 4 | 182 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCC62414.1 | 0.0 | 259 | 1735 | 28 | 1571 |
| QQA11075.1 | 6.84e-181 | 236 | 1299 | 92 | 1166 |
| BAB80970.1 | 9.46e-181 | 245 | 1299 | 110 | 1166 |
| AOY54034.1 | 2.50e-180 | 236 | 1299 | 92 | 1166 |
| ABG83191.1 | 4.78e-180 | 236 | 1299 | 92 | 1166 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7Q1W_A | 1.32e-07 | 1460 | 1634 | 69 | 271 | ChainA, Ruminococcus gnavus end galactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 7Q20_A | 1.32e-07 | 1460 | 1634 | 70 | 272 | ChainA, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149],7Q20_G Chain G, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 7PMO_D | 1.33e-07 | 1460 | 1634 | 70 | 272 | ChainD, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149],7PMO_G Chain G, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 2Y5P_A | 6.25e-06 | 1664 | 1725 | 6 | 68 | B-repeatof Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_B B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_C B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_D B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
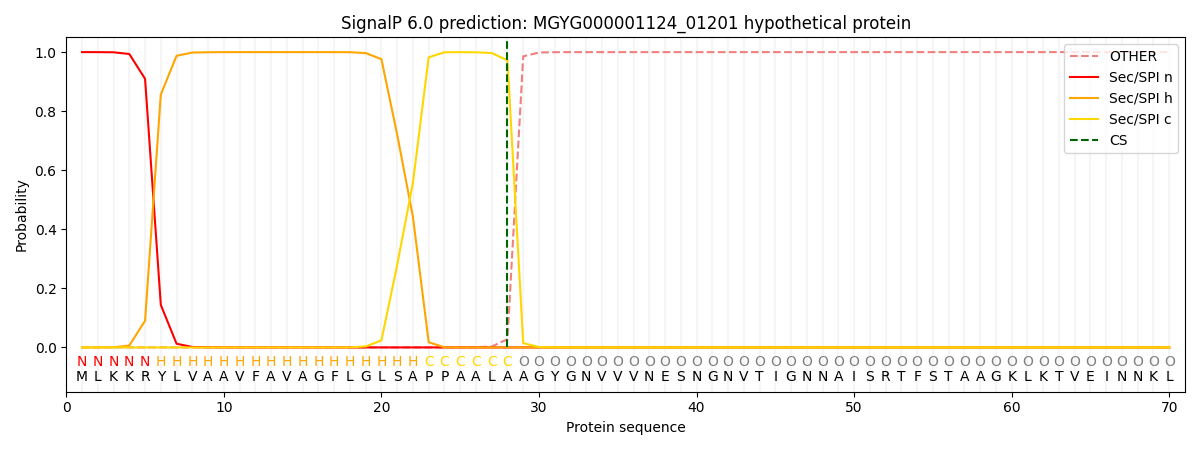
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998823 | 0.000195 | 0.000224 | 0.000206 | 0.000182 |