You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001129_00236
You are here: Home > Sequence: MGYG000001129_00236
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp000437255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp000437255 | |||||||||||
| CAZyme ID | MGYG000001129_00236 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45346; End: 47148 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 163 | 375 | 9.9e-18 | 0.8391304347826087 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1216 | GT2 | 1.65e-11 | 167 | 459 | 9 | 291 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
| pfam13641 | Glyco_tranf_2_3 | 2.55e-07 | 160 | 375 | 1 | 197 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUI96744.1 | 4.64e-140 | 3 | 596 | 5 | 614 |
| QOX65077.1 | 2.09e-100 | 6 | 597 | 7 | 611 |
| BAZ80590.1 | 5.87e-96 | 3 | 597 | 26 | 628 |
| BAY06970.1 | 7.04e-95 | 3 | 599 | 21 | 616 |
| AGA70580.1 | 8.16e-93 | 1 | 472 | 1 | 468 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FIX_A | 1.13e-28 | 49 | 456 | 75 | 490 | CrystalStructure of GlfT2 [Mycobacterium tuberculosis H37Rv],4FIX_B Crystal Structure of GlfT2 [Mycobacterium tuberculosis H37Rv],4FIY_A Crystal Structure of GlfT2 Complexed with UDP [Mycobacterium tuberculosis H37Rv],4FIY_B Crystal Structure of GlfT2 Complexed with UDP [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0R628 | 7.27e-31 | 49 | 456 | 66 | 479 | Galactofuranosyltransferase GlfT2 OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=glfT2 PE=3 SV=1 |
| O53585 | 5.69e-28 | 49 | 456 | 55 | 470 | Galactofuranosyltransferase GlfT2 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=glfT2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
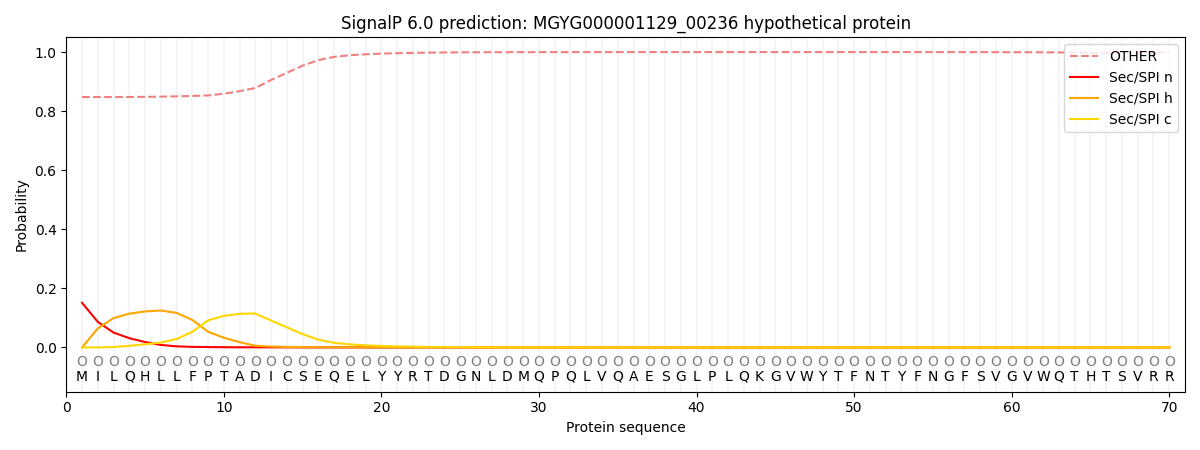
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.857512 | 0.139976 | 0.001456 | 0.000417 | 0.000207 | 0.000445 |
