You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001151_01856
You are here: Home > Sequence: MGYG000001151_01856
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
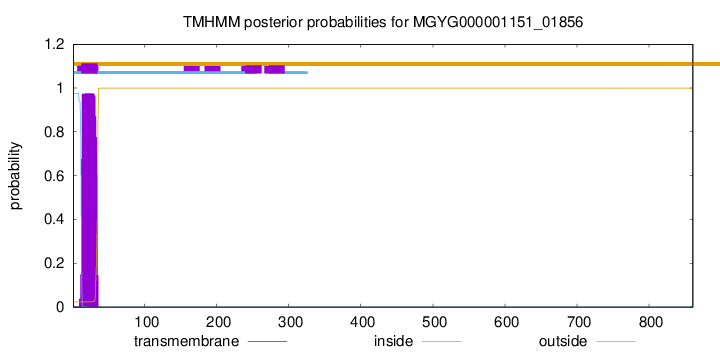
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp000433635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp000433635 | |||||||||||
| CAZyme ID | MGYG000001151_01856 | |||||||||||
| CAZy Family | CBM79 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30283; End: 32868 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18522 | DUF5620 | 4.01e-22 | 513 | 649 | 1 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| pfam18522 | DUF5620 | 5.27e-22 | 298 | 490 | 1 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| pfam18522 | DUF5620 | 5.26e-16 | 79 | 179 | 20 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| cd14256 | Dockerin_I | 1.33e-11 | 789 | 846 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 1.25e-05 | 790 | 846 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCZ83178.1 | 0.0 | 1 | 861 | 1 | 877 |
| EWM54821.1 | 2.11e-47 | 219 | 600 | 185 | 511 |
| CBL17555.1 | 5.24e-42 | 7 | 650 | 8 | 506 |
| CCZ84116.1 | 1.38e-08 | 789 | 848 | 747 | 808 |
| ERJ92224.1 | 8.45e-07 | 789 | 852 | 784 | 846 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
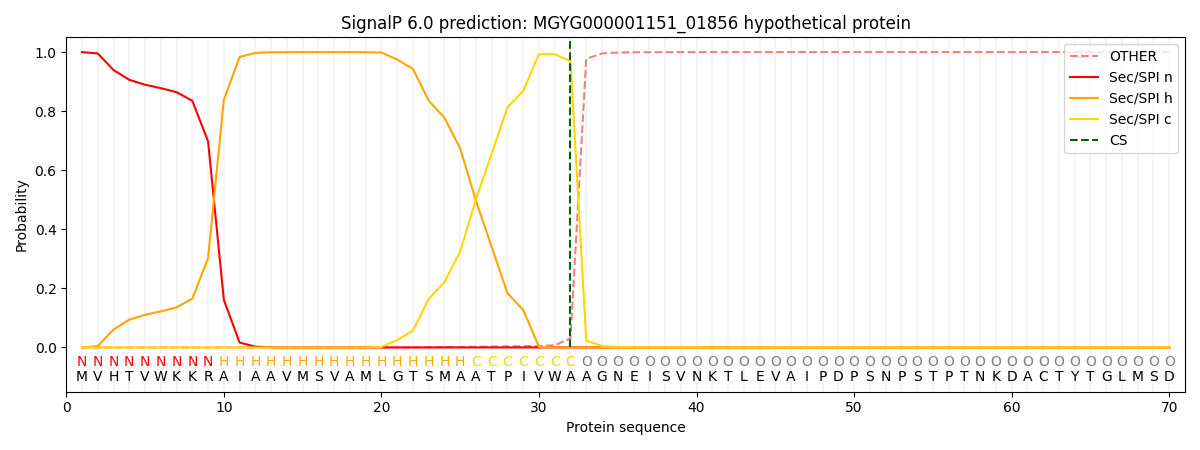
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.999020 | 0.000157 | 0.000198 | 0.000166 | 0.000141 |