You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001151_02242
You are here: Home > Sequence: MGYG000001151_02242
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
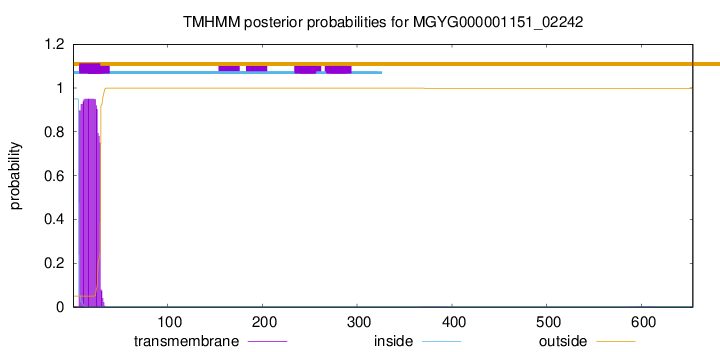
TMHMM annotations
Basic Information help
| Species | Ruminococcus_C sp000433635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; Ruminococcus_C sp000433635 | |||||||||||
| CAZyme ID | MGYG000001151_02242 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 335; End: 2299 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 39 | 190 | 3.3e-28 | 0.654639175257732 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01833 | XynB_like | 2.00e-30 | 39 | 279 | 1 | 155 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 8.37e-16 | 44 | 271 | 2 | 175 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam00657 | Lipase_GDSL | 1.91e-14 | 41 | 277 | 1 | 224 | GDSL-like Lipase/Acylhydrolase. |
| cd00229 | SGNH_hydrolase | 8.92e-11 | 41 | 280 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd01832 | SGNH_hydrolase_like_1 | 1.80e-08 | 135 | 281 | 64 | 182 | Members of the SGNH-hydrolase superfamily, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. Myxobacterial members of this subfamily have been reported to be involved in adventurous gliding motility. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAB55348.1 | 2.12e-49 | 20 | 286 | 22 | 265 |
| AWI67000.1 | 1.15e-43 | 34 | 283 | 92 | 316 |
| AAQ10005.1 | 3.50e-39 | 37 | 291 | 43 | 269 |
| AAQ10006.1 | 3.50e-39 | 37 | 291 | 43 | 269 |
| AAB69092.1 | 6.52e-36 | 34 | 288 | 58 | 282 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VPT_A | 9.92e-14 | 39 | 286 | 6 | 200 | ChainA, LIPOLYTIC ENZYME [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RLB8 | 4.25e-50 | 20 | 286 | 22 | 265 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
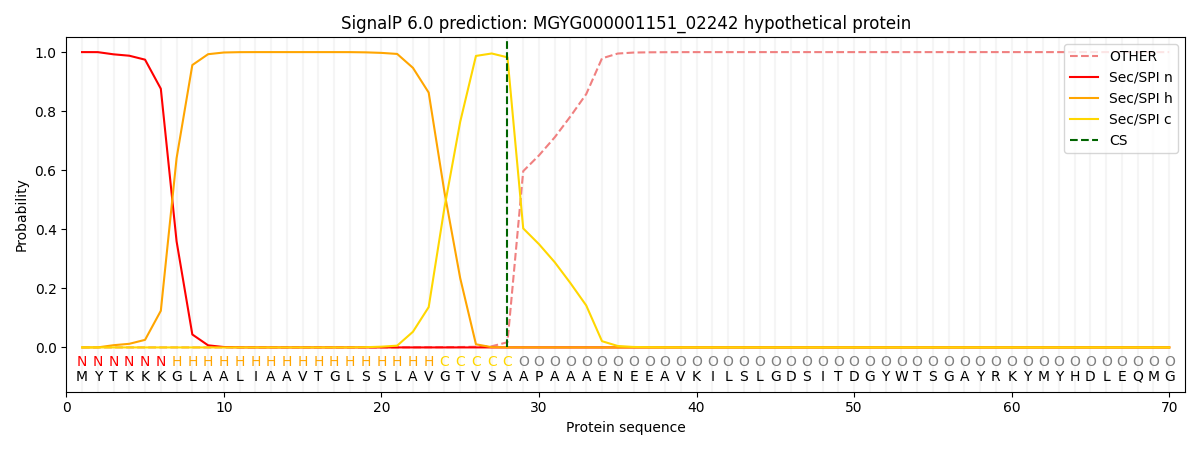
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000269 | 0.998989 | 0.000178 | 0.000218 | 0.000190 | 0.000159 |