You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001159_01421
Basic Information
help
| Species |
Bifidobacterium pullorum
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium pullorum
|
| CAZyme ID |
MGYG000001159_01421
|
| CAZy Family |
GT2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1016 |
|
108759.75 |
7.8263 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001159 |
2232439 |
MAG |
Austria |
Europe |
|
| Gene Location |
Start: 57127;
End: 60177
Strand: +
|
No EC number prediction in MGYG000001159_01421.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1216
|
GT2 |
6.32e-07 |
109 |
251 |
76 |
219 |
Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000021
|
0.000021
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
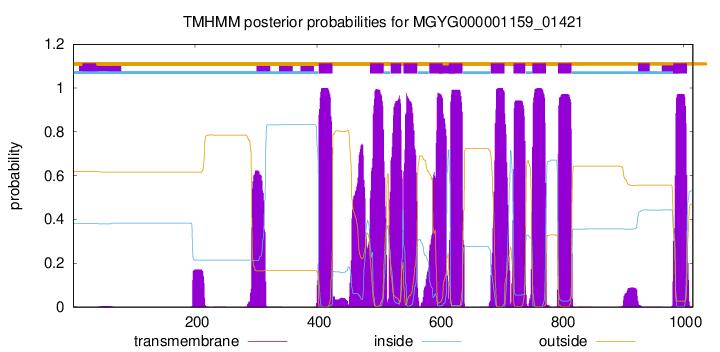
| start |
end |
| 403 |
425 |
| 487 |
509 |
| 521 |
538 |
| 542 |
564 |
| 584 |
606 |
| 616 |
638 |
| 685 |
707 |
| 722 |
741 |
| 753 |
775 |
| 795 |
817 |
| 984 |
1006 |


