You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001164_02164
You are here: Home > Sequence: MGYG000001164_02164
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella lascolaii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella lascolaii | |||||||||||
| CAZyme ID | MGYG000001164_02164 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3064; End: 7515 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 870 | 1347 | 1.6e-102 | 0.5492021276595744 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16335 | DUF4965 | 1.25e-89 | 487 | 656 | 1 | 174 | Domain of unknown function (DUF4965). This family consists of uncharacterized proteins around 840 residues in length and is mainly found in various Bacteroides species. Several proteins in this family are annotated as Glutaminases, but the function of this protein is unknown. |
| pfam08760 | DUF1793 | 1.47e-75 | 662 | 827 | 1 | 169 | Domain of unknown function (DUF1793). This presumed domain is found at the C-terminus of a glutaminase protein from fungi. This domain is also found as a single domain protein in Bacteroides thetaiotaomicron. |
| pfam17168 | DUF5127 | 5.68e-69 | 259 | 483 | 1 | 226 | Domain of unknown function (DUF5127). |
| COG3250 | LacZ | 1.06e-35 | 878 | 1353 | 15 | 481 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.93e-35 | 877 | 1274 | 14 | 418 | beta-D-glucuronidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA48091.1 | 4.25e-310 | 140 | 830 | 4 | 700 |
| AHW60263.1 | 7.23e-210 | 838 | 1431 | 18 | 601 |
| BBE17791.1 | 1.24e-209 | 840 | 1431 | 7 | 588 |
| QIA07412.1 | 1.69e-207 | 838 | 1431 | 18 | 601 |
| SNV38566.1 | 3.28e-206 | 840 | 1431 | 20 | 600 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 3.24e-188 | 840 | 1431 | 4 | 586 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 4JKM_A | 2.11e-32 | 869 | 1273 | 9 | 415 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 5UJ6_A | 1.37e-27 | 926 | 1273 | 79 | 418 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 1.39e-27 | 926 | 1273 | 87 | 426 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 6D8G_A | 1.24e-26 | 926 | 1273 | 87 | 426 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2U4L7 | 2.03e-73 | 248 | 821 | 94 | 683 | Glutaminase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=gtaA PE=1 SV=2 |
| D4AMT2 | 1.11e-42 | 482 | 830 | 350 | 786 | Probable glutaminase ARB_05535/05536 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_05535/05536 PE=1 SV=2 |
| T2KPJ7 | 1.16e-26 | 916 | 1275 | 96 | 432 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| A7LXS9 | 1.62e-21 | 869 | 1336 | 38 | 544 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| P06760 | 1.66e-20 | 878 | 1309 | 41 | 488 | Beta-glucuronidase OS=Rattus norvegicus OX=10116 GN=Gusb PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
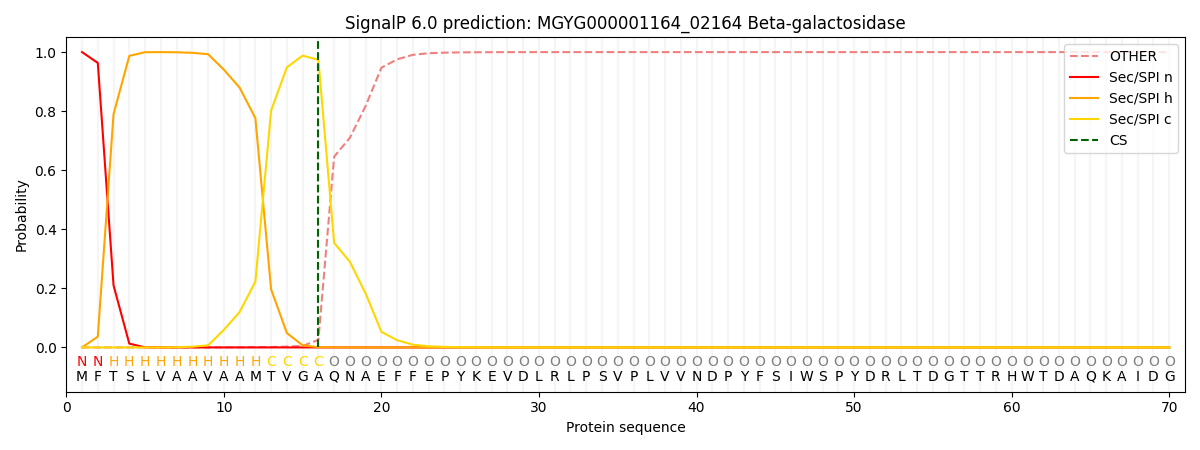
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000373 | 0.998910 | 0.000154 | 0.000205 | 0.000168 | 0.000155 |
