You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001180_01027
You are here: Home > Sequence: MGYG000001180_01027
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp001701195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp001701195 | |||||||||||
| CAZyme ID | MGYG000001180_01027 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66263; End: 69754 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 749 | 987 | 5.5e-37 | 0.8576388888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 7.48e-09 | 751 | 1028 | 16 | 296 | Pectinesterase. |
| COG4677 | PemB | 8.28e-09 | 745 | 961 | 89 | 348 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02488 | PLN02488 | 1.84e-08 | 744 | 950 | 196 | 419 | probable pectinesterase/pectinesterase inhibitor |
| PLN02432 | PLN02432 | 1.17e-07 | 829 | 961 | 101 | 227 | putative pectinesterase |
| PLN02217 | PLN02217 | 2.07e-06 | 791 | 959 | 301 | 481 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37302.1 | 1.24e-274 | 29 | 1163 | 31 | 1159 |
| ANU64595.1 | 1.24e-274 | 29 | 1163 | 31 | 1159 |
| QQR08039.1 | 1.24e-274 | 29 | 1163 | 31 | 1159 |
| QUT74167.1 | 4.18e-65 | 586 | 1154 | 866 | 1425 |
| QCD38476.1 | 4.60e-60 | 642 | 1162 | 934 | 1474 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 1.03e-11 | 761 | 948 | 53 | 269 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NPS7 | 9.90e-09 | 753 | 994 | 50 | 295 | Probable pectinesterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pmeA PE=3 SV=1 |
| Q2UBD9 | 9.90e-09 | 753 | 994 | 50 | 295 | Probable pectinesterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pmeA PE=3 SV=1 |
| O48711 | 2.87e-08 | 769 | 1030 | 266 | 539 | Probable pectinesterase/pectinesterase inhibitor 12 OS=Arabidopsis thaliana OX=3702 GN=PME12 PE=2 SV=1 |
| Q8RXK7 | 3.92e-08 | 756 | 1027 | 275 | 554 | Probable pectinesterase/pectinesterase inhibitor 41 OS=Arabidopsis thaliana OX=3702 GN=PME41 PE=2 SV=2 |
| Q9SRX4 | 3.55e-07 | 753 | 1023 | 280 | 556 | Probable pectinesterase/pectinesterase inhibitor 7 OS=Arabidopsis thaliana OX=3702 GN=PME7 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
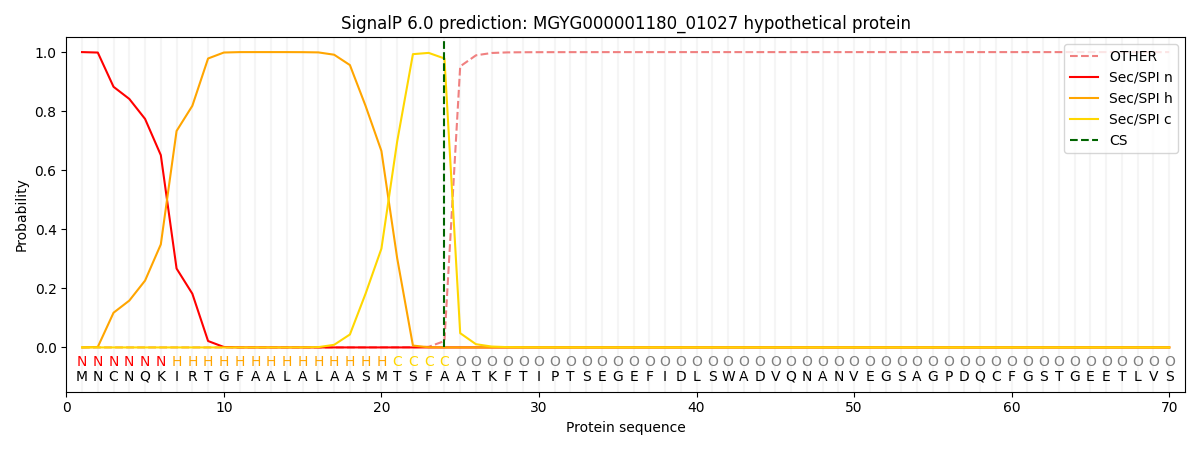
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000302 | 0.999039 | 0.000174 | 0.000166 | 0.000158 | 0.000142 |
