You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001183_00356
You are here: Home > Sequence: MGYG000001183_00356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900551755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900551755 | |||||||||||
| CAZyme ID | MGYG000001183_00356 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 367940; End: 369523 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 251 | 487 | 9.8e-92 | 0.9915611814345991 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.74e-70 | 248 | 494 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.32e-13 | 233 | 489 | 40 | 276 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00395 | SLH | 7.52e-10 | 38 | 79 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 3.43e-04 | 29 | 110 | 634 | 721 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 0.001 | 175 | 200 | 17 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT51240.1 | 4.45e-217 | 38 | 526 | 27 | 515 |
| QQR30621.1 | 7.19e-192 | 38 | 526 | 32 | 519 |
| ANU55408.1 | 7.19e-192 | 38 | 526 | 32 | 519 |
| ASB41357.1 | 7.19e-192 | 38 | 526 | 32 | 519 |
| CUH91792.1 | 3.42e-111 | 238 | 527 | 41 | 331 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 3.27e-110 | 234 | 522 | 7 | 296 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 9.46e-105 | 219 | 522 | 16 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 8.49e-102 | 234 | 522 | 6 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 6.98e-101 | 234 | 522 | 6 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1A3H_A | 2.78e-99 | 234 | 522 | 3 | 293 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15704 | 3.53e-111 | 224 | 522 | 31 | 331 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P07983 | 2.44e-100 | 234 | 522 | 36 | 325 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 3.44e-100 | 234 | 522 | 36 | 325 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P23549 | 1.18e-97 | 234 | 522 | 36 | 325 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
| O85465 | 3.78e-97 | 234 | 522 | 32 | 322 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
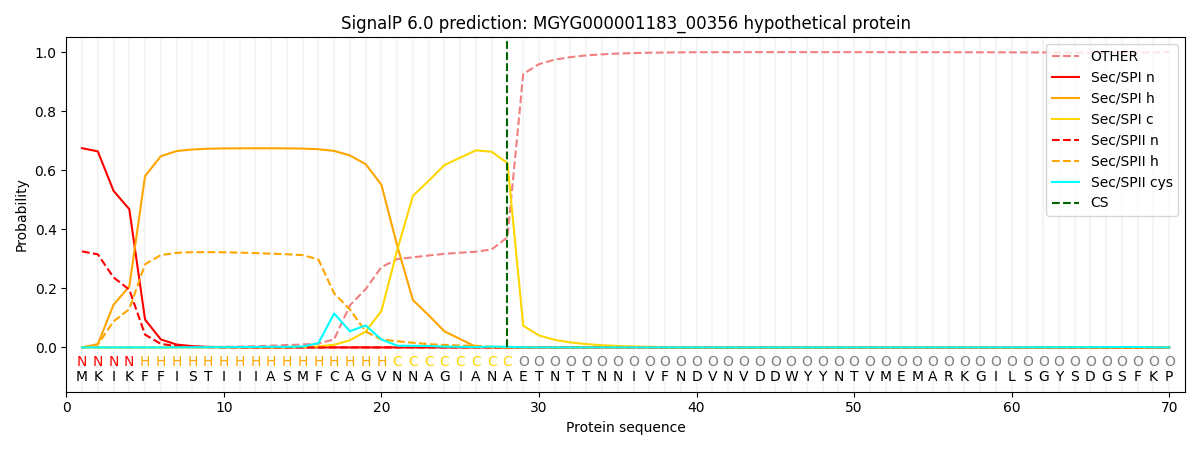
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000907 | 0.664942 | 0.333231 | 0.000381 | 0.000279 | 0.000228 |
